Preprints

J Kreger, E Gonzalez, X Wu, ET Roussos Torres, AL MacLean
bioRxiv (2025); 10.1101/2025.08.11.669777. bioRxiv
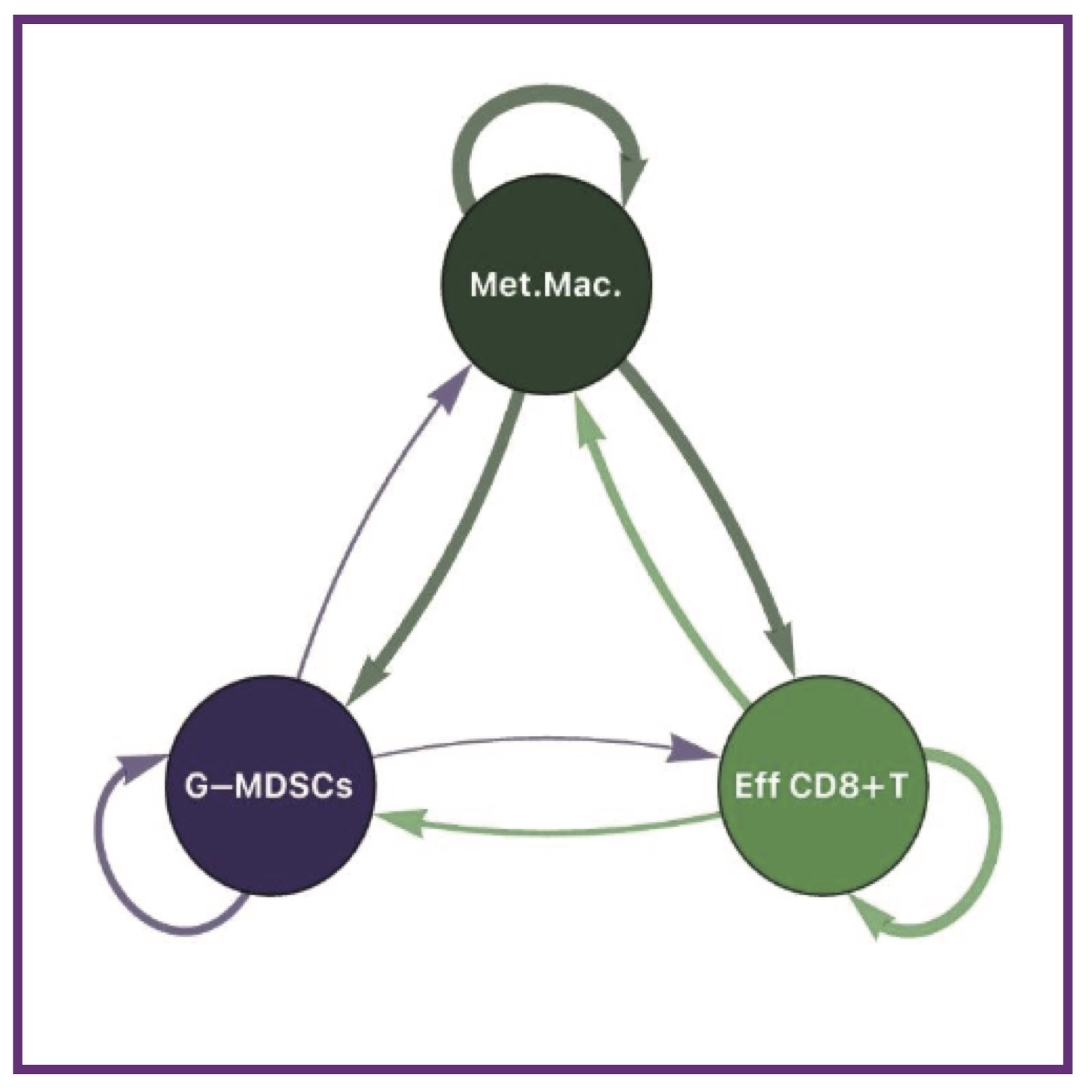
E Gonzalez, J Kreger, Y Liu, X Wu, A Barbetta, AG Baugh, B Al-Zubeidy, J Jang, SM Shin, M Jacobo, V Stearns, RM Connolly, W Ho, J Emamaullee, AL MacLean, ET Roussos Torres
bioRxiv (2025); 10.1101/2025.06.09.658361. bioRxiv
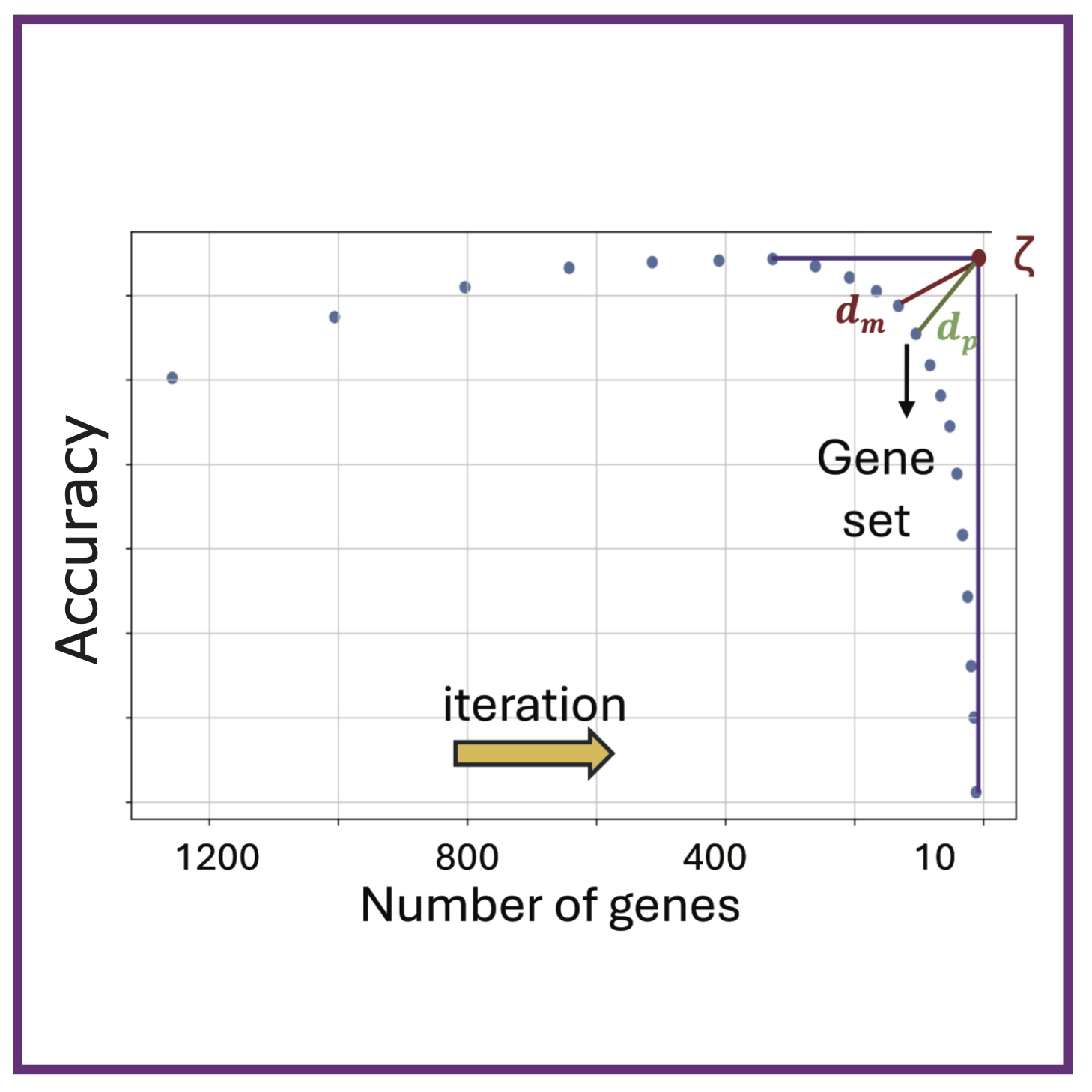
Y Liu, A Baugh, ET Roussos Torres, AL MacLean
bioRxiv (2025); 10.1101/2025.05.23.655858. bioRxiv
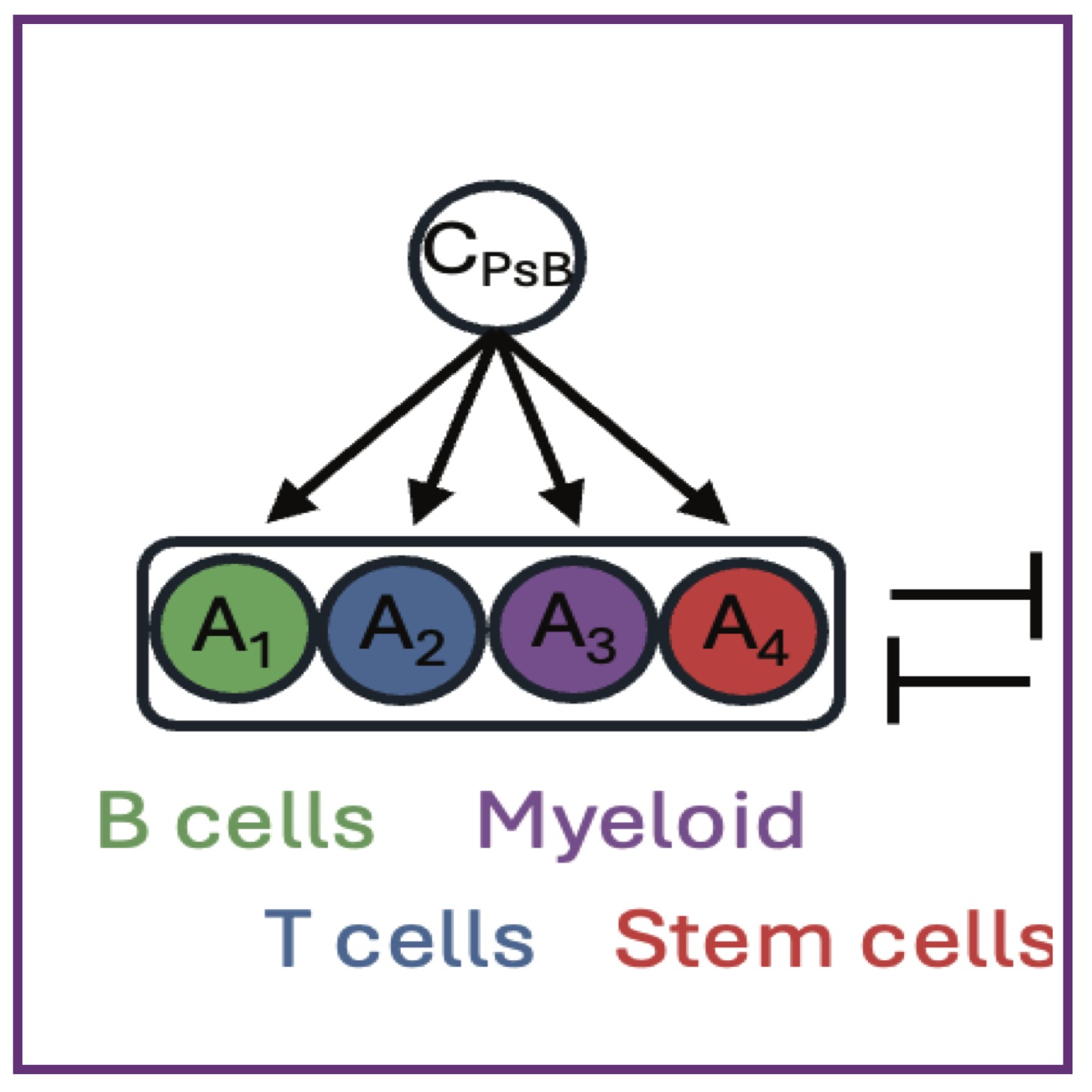
DE Frankhouser, D Zhao, Y Fu, A Dey, Z Chen, J Irizarry, JR Ambriz, S Branciamore, D O’Meally, L Ghoda, JM Trent, S Forman, AL MacLean, Y Kuo, B Zhang, RC Rockne, G Marcucci
bioRxiv (2025); 10.1101/2025.05.14.653262. bioRxiv
Publications
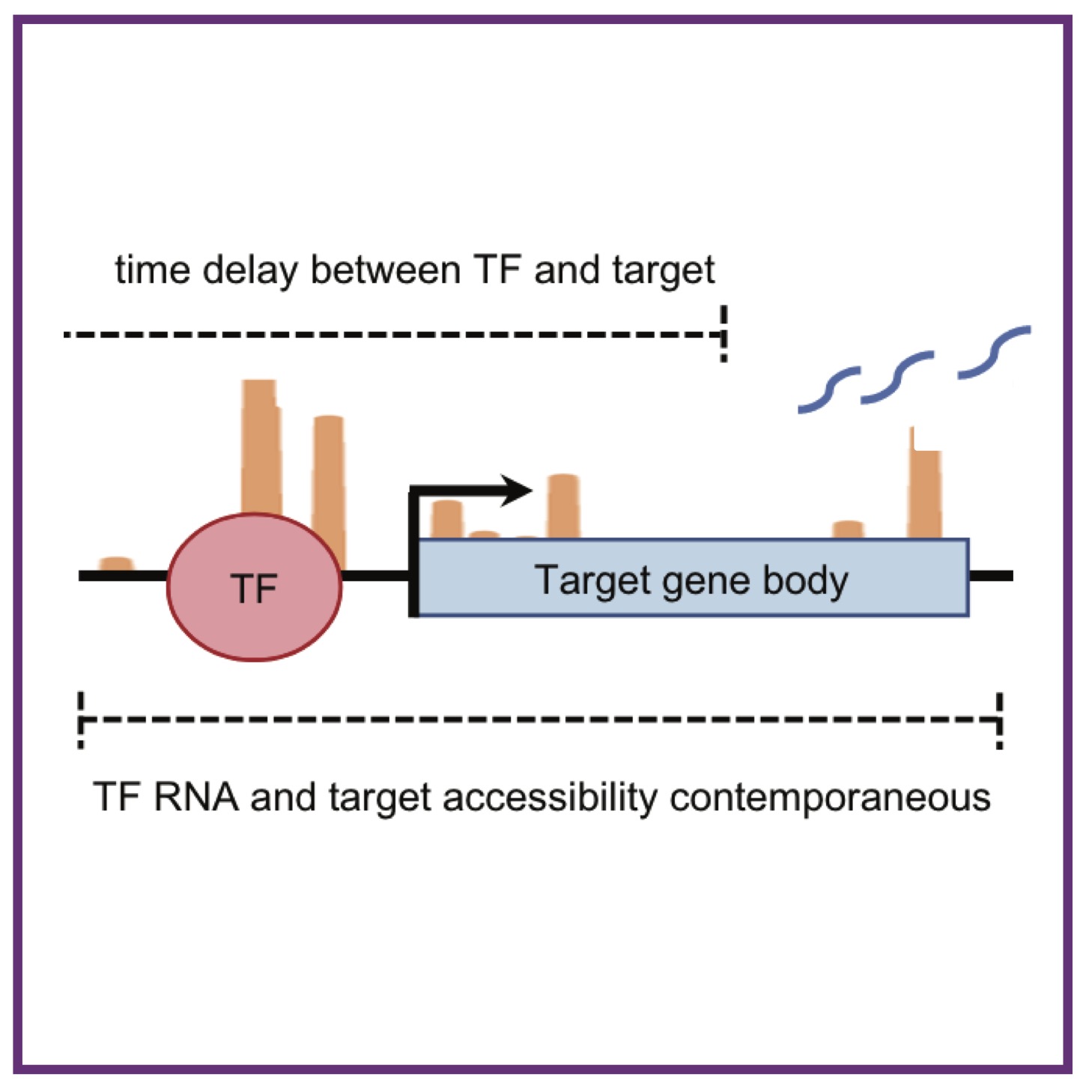
MK Rommelfanger, M Behrends, Y Chen, J Martinez, N Kurella, N Geisler, D Guturu, M Bens, L Xiong, Z Xiang, KL Rudolph, AL MacLean
iScience (2025); 10.1016/j.isci.2025.114010. iScience

A Dey, AL MacLean
Development (2025); 10.1242/dev.204583. PubMed Development
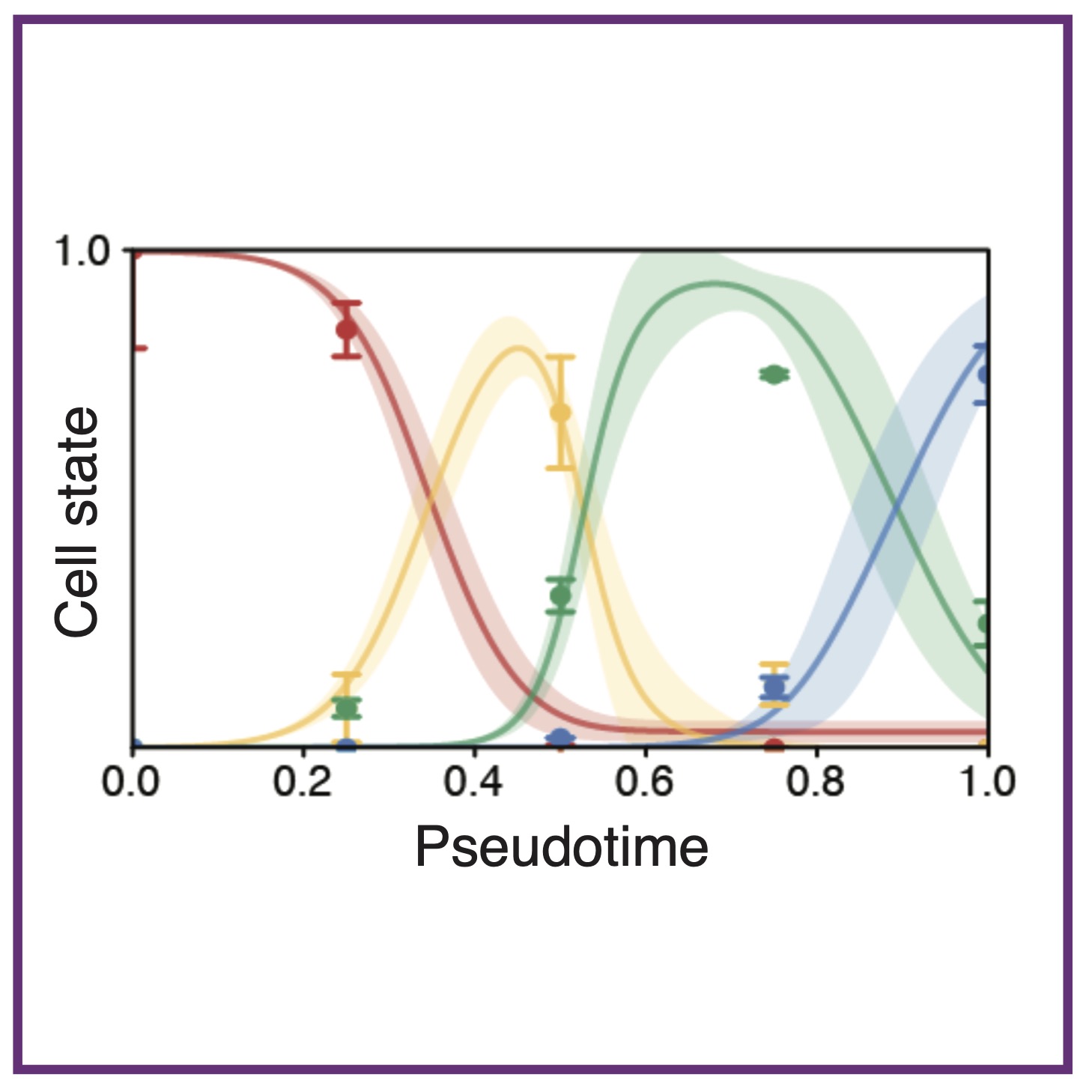
M McDermott, R Mehta, ET Roussos Torres, AL MacLean
npj Syst Biol Appl (2025); 10.1038/s41540-025-00512-2. PubMed npj Syst Biol Appl
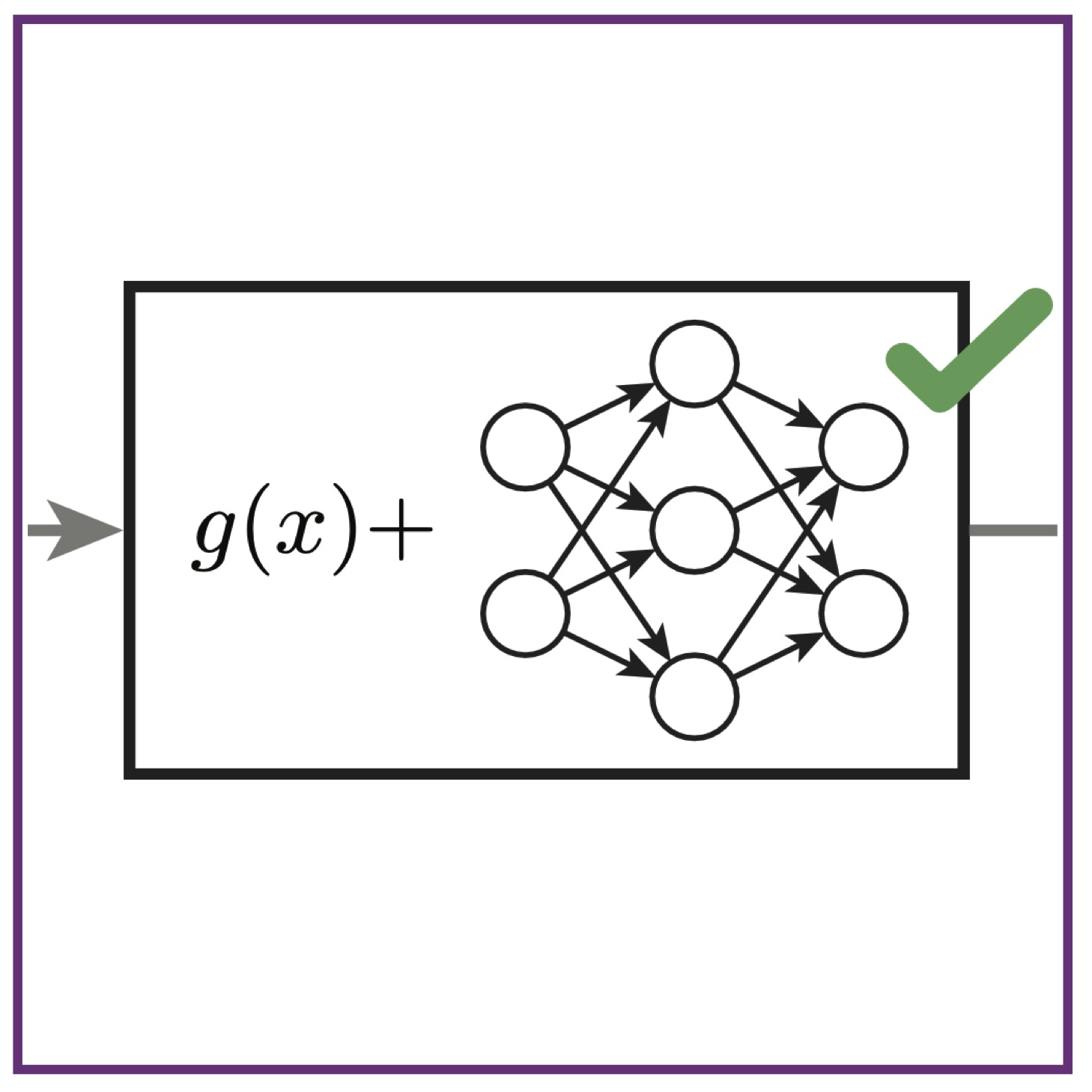
X Wu, M McDermott, AL MacLean
PLOS Comp Biol (2025); 10.1371/journal.pcbi.1012762. PubMed PLOS Comp Biol
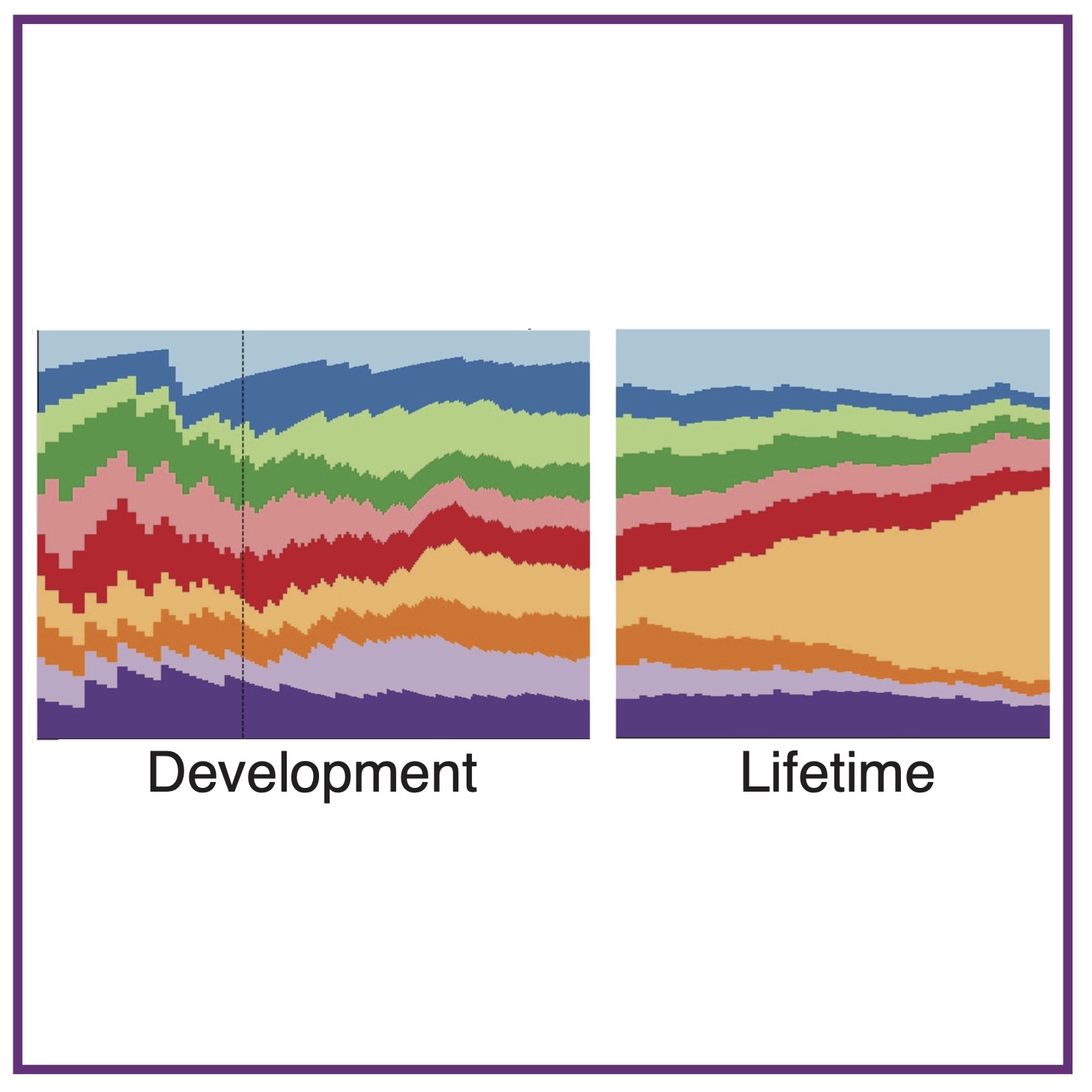
J Kreger, JA Mooney, D Shibata, AL MacLean
Nat Commun (2024); 10.1038/s41467-024-54711-2 PubMed Nat Commun
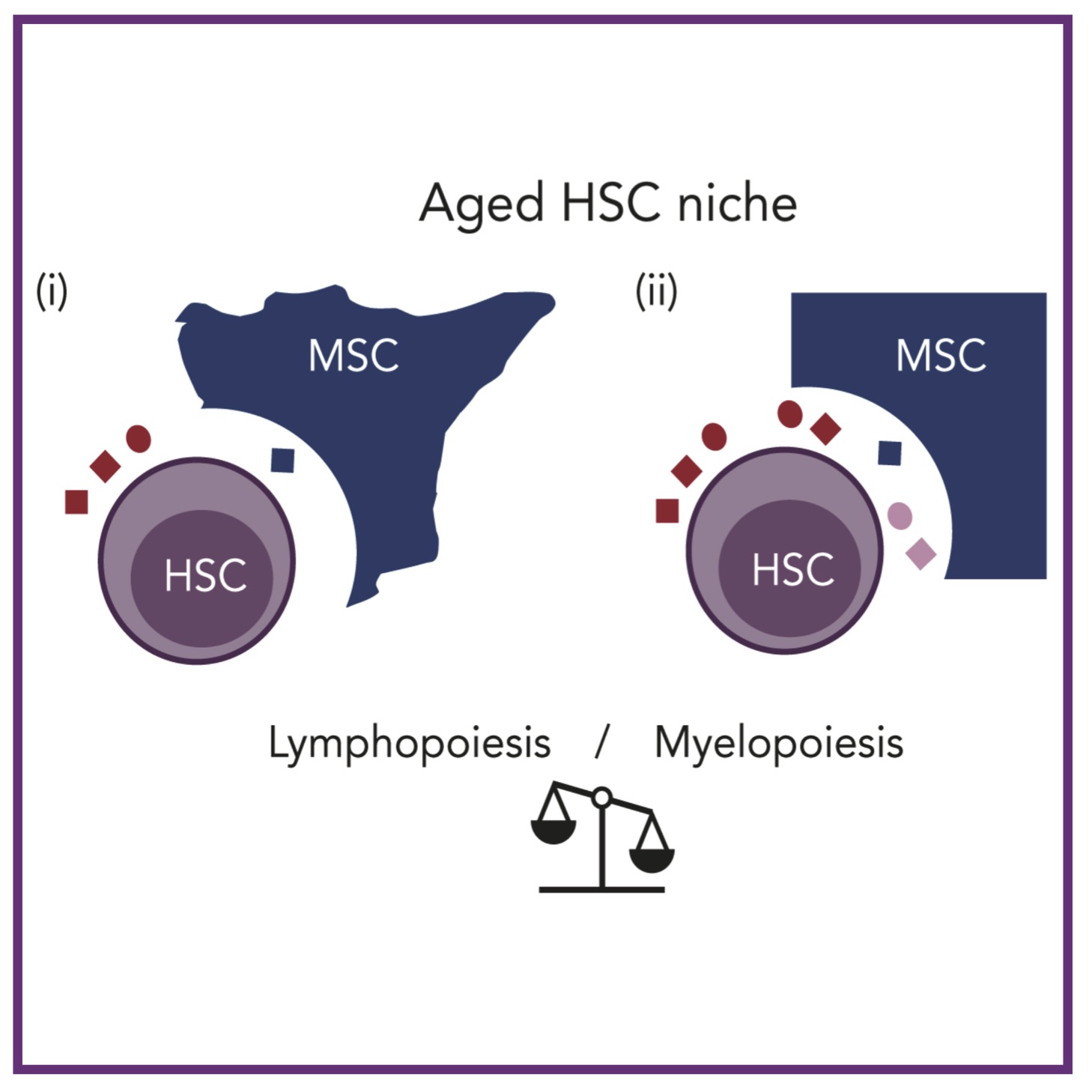
AL MacLean, KL Rudolph
Blood (2024); 10.1182/blood.2024024890 PubMed Blood

JL Stellman, AE Zambidis, AL MacLean, DJ Kast
Connected Science Learning (2024); 10.1080/24758779.2024.2439370. Conn Sci Learn
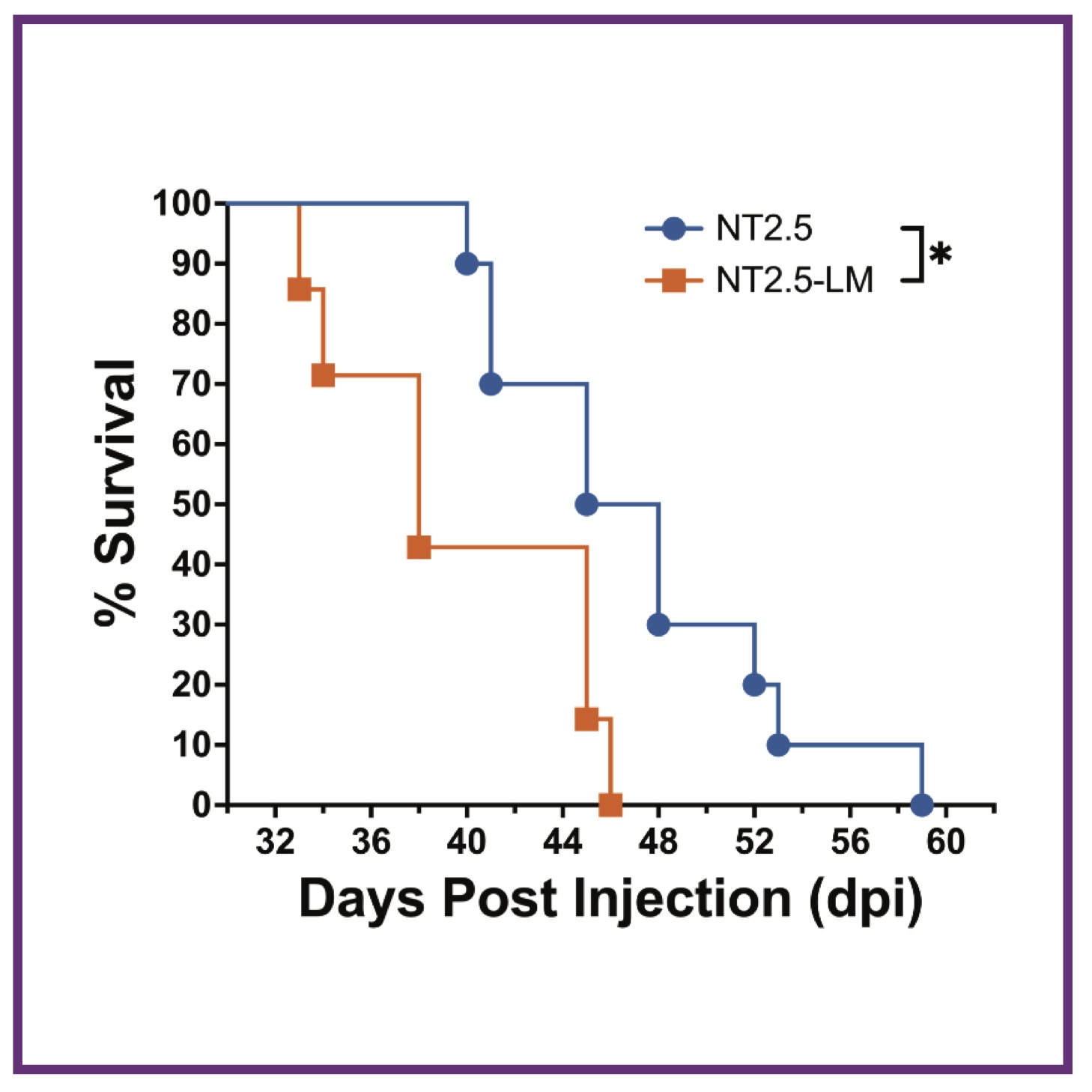
AG Baugh, E Gonzalez, VH Narumi, J Kreger, Y Liu, C Rafie, S Castanon, J Jang, LT Kagohara, DP Anastasiadou, J Leatherman, TD Armstrong, I Chan, GS Karagiannis, EM Jaffee, AL MacLean, ET Roussos Torres
Clin Exp Metas (2024); 10.1007/s10585-024-10289-z PubMed Clin Exp Metas
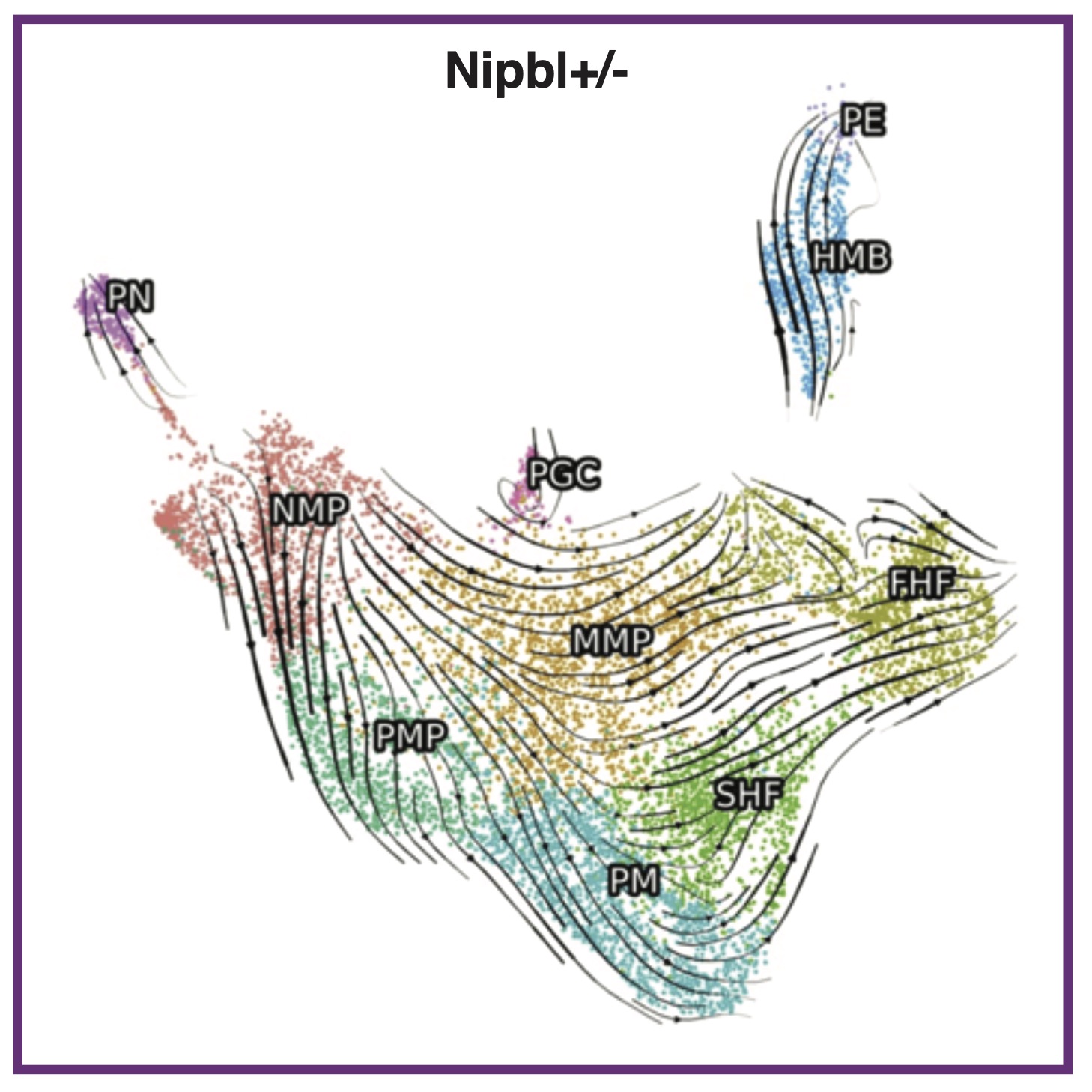
S Chea, J Kreger, ME Lopez-Burks, AL MacLean, AD Lander, AL Calof
Science Advances (2024); 10.1126/sciadv.add9984 PubMed Science Advances
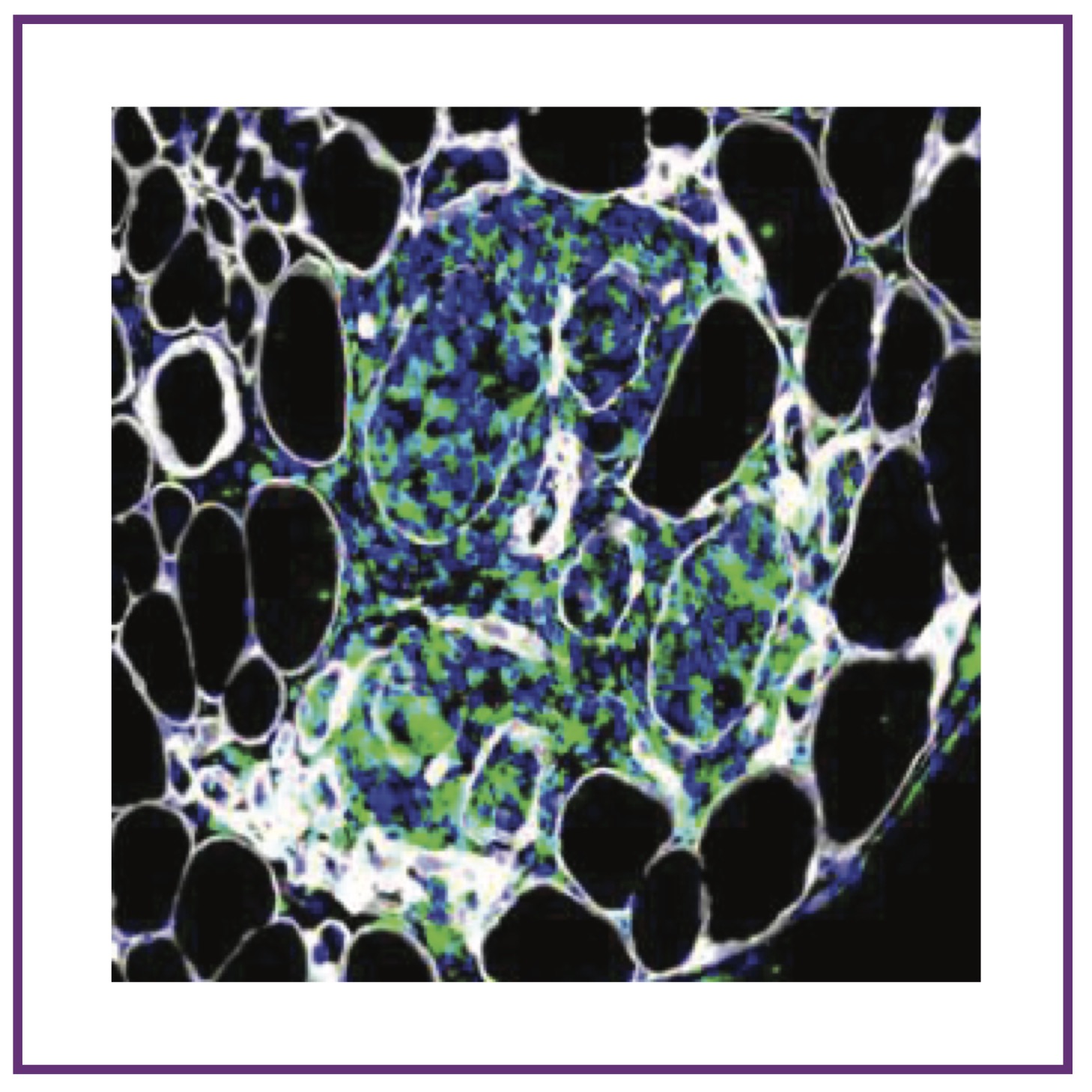
G Coulis et al.
Science Advances (2023); 10.1126/sciadv.add9984 PubMed Science Advances
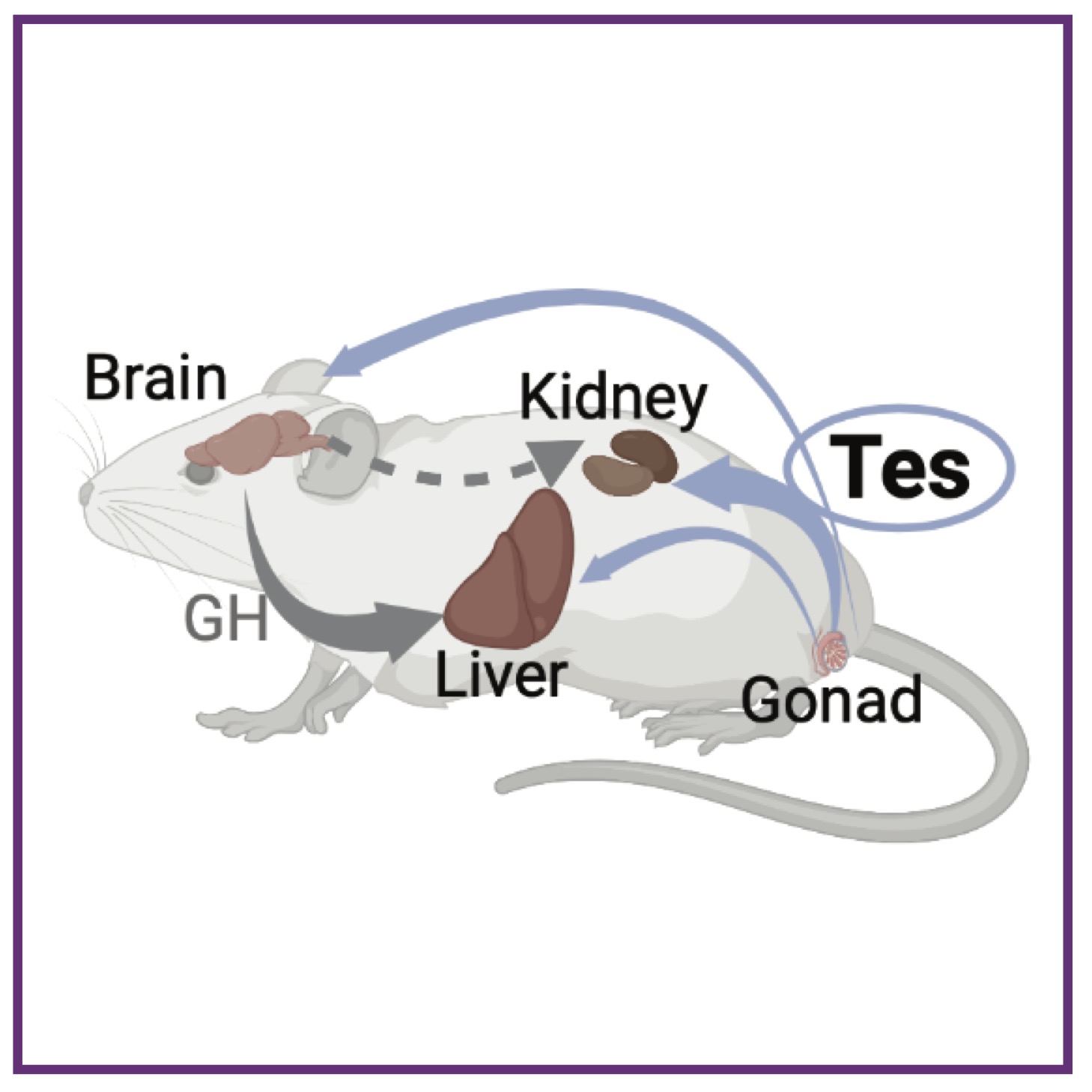
L Xiong, J Liu, SY Han, K Koppitch, JJ Guo, MK Rommelfanger, F Gao, IB Hallgrimsdottir, L Pachter, J Kim, AL MacLean, AP McMahon
Dev Cell (2023); 10.1016/j.devcel.2023.08.010 PubMed Dev Cell
X Wu, R Wollman, AL MacLean
J R Soc Interface (2023); 10.1098/rsif.2023.0172 PubMed J R Soc Interface
J Kreger, ET Roussos Torres, AL MacLean
Can Immunol Res (2023); 10.1158/2326-6066.CIR-22-0617 PubMed Can Immunol Res
E Roesch, JG Greener, AL MacLean, H Nassar, C Rackauckas, TE Holy, MPH Stumpf
Nat Meth (2023); 10.1038/s41592-023-01832-z PubMed Nat Meth
AL MacLean
Cell Rep Meth (2022); 10.1016/j.crmeth.2022.100204 PubMed Cell Rep Meth
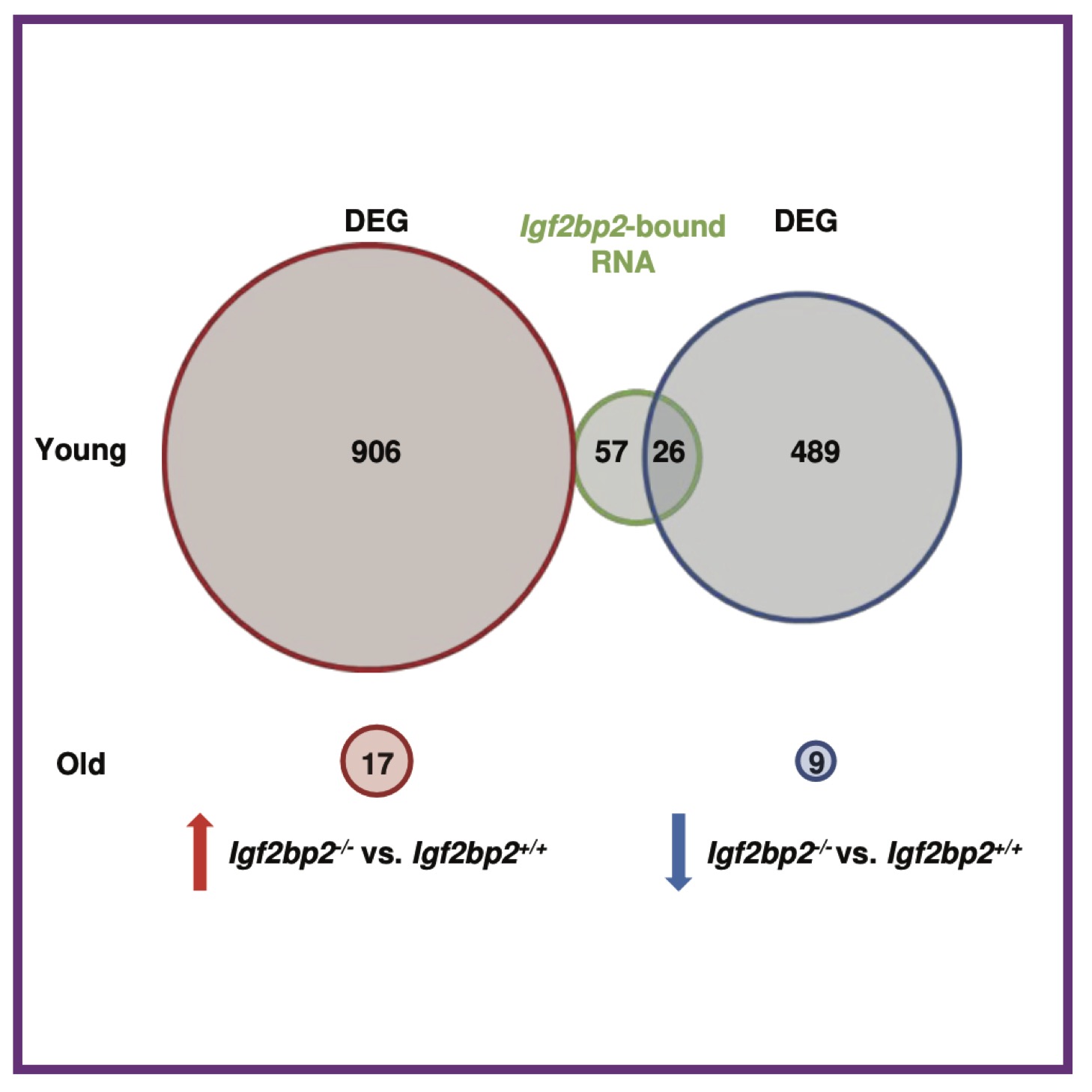
M Suo, MK Rommelfanger, Y Chen, EM Amro, B Han, Z Chen, K Szafranski, SR Chakkarappan, BO Boehm, AL MacLean, KL Rudolph
Blood (2022); doi.org/10.1182/blood.2021012197 PubMed Blood
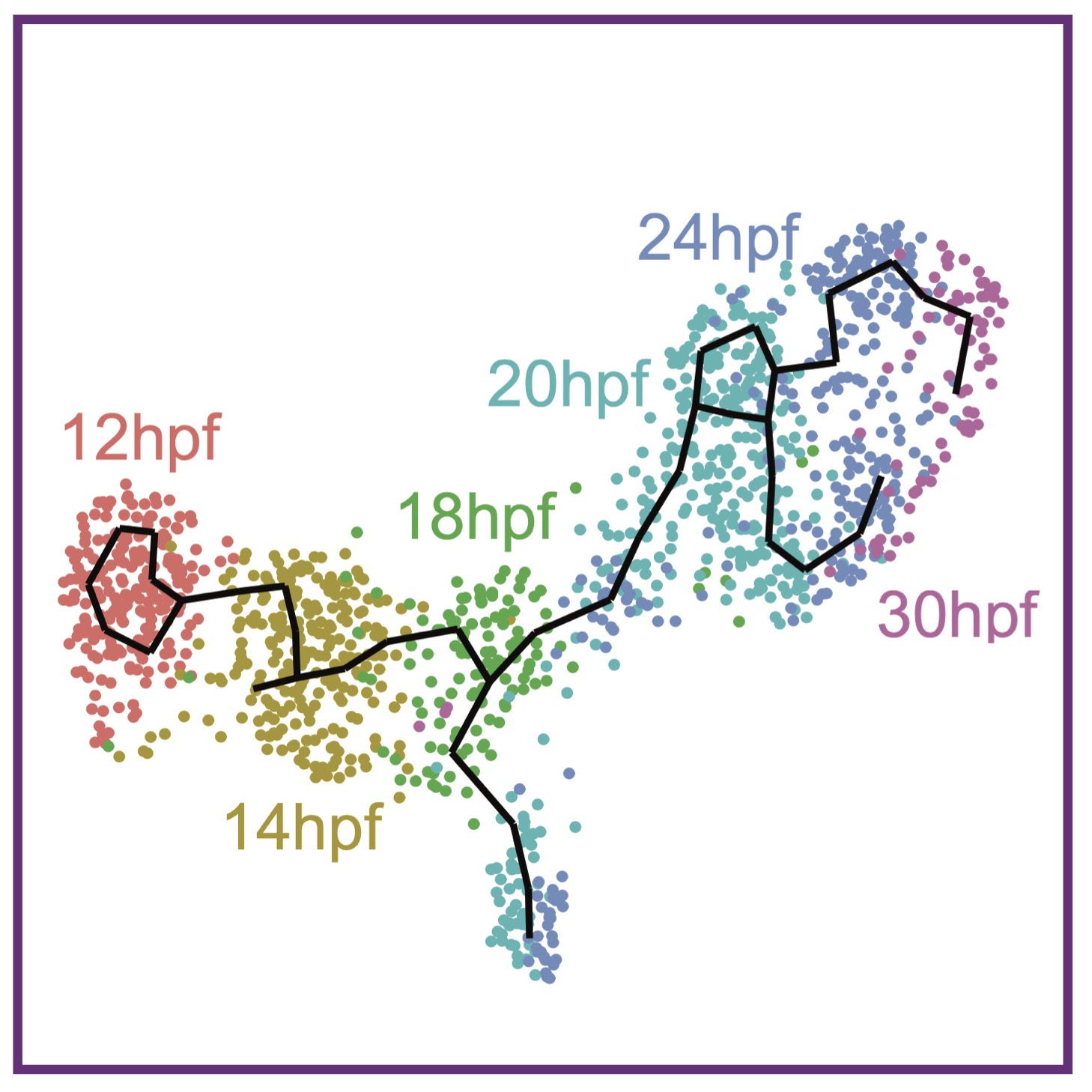
D Tatarakis, Z Cang, X Wu, P Sharma, MK Karikomi, AL MacLean, Q Nie, TF Schilling
Cell Reports (2021); 37(12), 110140 PubMed Cell Rep
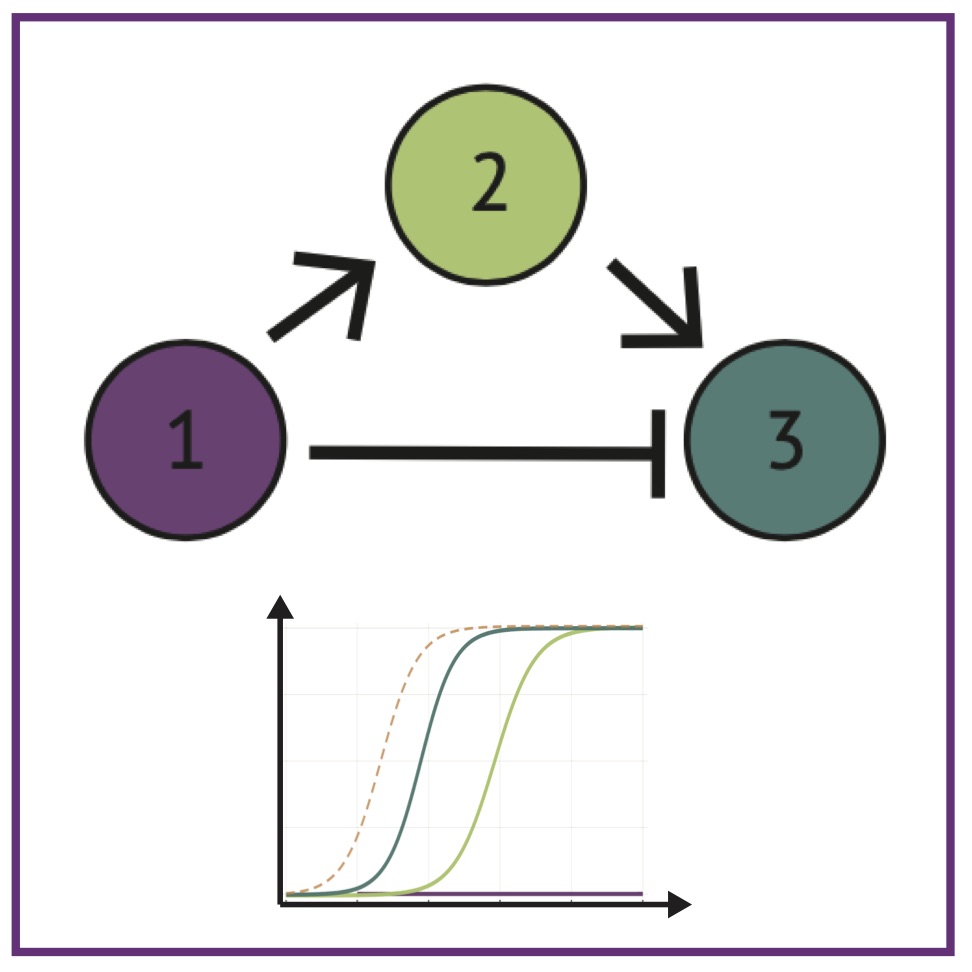
MK Rommelfanger, AL MacLean
Development (2021); 148(24), dev199779 PubMed Development
HM Rando et al.
mSystems (2021); 6(5) PubMed mSystems
AL MacLean, Q Nie
Phys Biol, (2021); 18:050201 PubMed Phys Biol
R Mitra, AL MacLean
Bioinformatics, (2021); btab260 PubMed Bioinformatics
DR Bergman, MK Karikomi, Q Nie, AL MacLean
Communications Biol, (2021); 4:983 PubMed Communications Biol
 Single cell transcriptomics of human epidermis reveals basal stem cell transition states
Single cell transcriptomics of human epidermis reveals basal stem cell transition statesS Wang, ML Drummond, CF Guerrero-Juarez, E Tarapore, AL MacLean, AR Stabell, SC Wu, G Gutierrez, BT That, CA Benavente, Q Nie, SX Atwood
S Wang et al.
Nat Commun, 2020; 11:4239 PubMed Nat Commun
 Cell lineage and communication network inference via optimization for single-cell transcriptomics
Cell lineage and communication network inference via optimization for single-cell transcriptomicsS Wang, MK Karikomi, AL MacLean, Q Nie
S Wang et al.
Nucleic Acids Res, 2019. gkz204 Pubmed Nucleic Acids Res
AL MacLean*, T Hong*, Q Nie
Curr Opin Sys Biol, 2018. 9:32-41 Curr Opin Sys Biol
 14. Energy landscape-based inference of transition probabilities and cellular trajectories from single-cell transcriptomic data
14. Energy landscape-based inference of transition probabilities and cellular trajectories from single-cell transcriptomic dataS Jin, AL MacLean, T Peng, Q Nie
Bioinformatics, 2018. 34(12):2077-2086 Pubmed Bioinformatics
 Bayesian inference of agent-based models: a tool for studying kidney branching morphogenesis
Bayesian inference of agent-based models: a tool for studying kidney branching morphogenesisBen Lambert*, AL MacLean*, AG Fletcher, AN Coombes, MH Little, HM Byrne
Ben Lambert*, AL MacLean*, AG Fletcher, et al.
J Math Biol, 2018. 76(7):1673-1697 Pubmed J Math Biol
Y Guo, Q Nie, AL MacLean, Y Li, J Lei, and S Li
Cancer Research, 2017. 77(22):6429–41 Pubmed Cancer Res
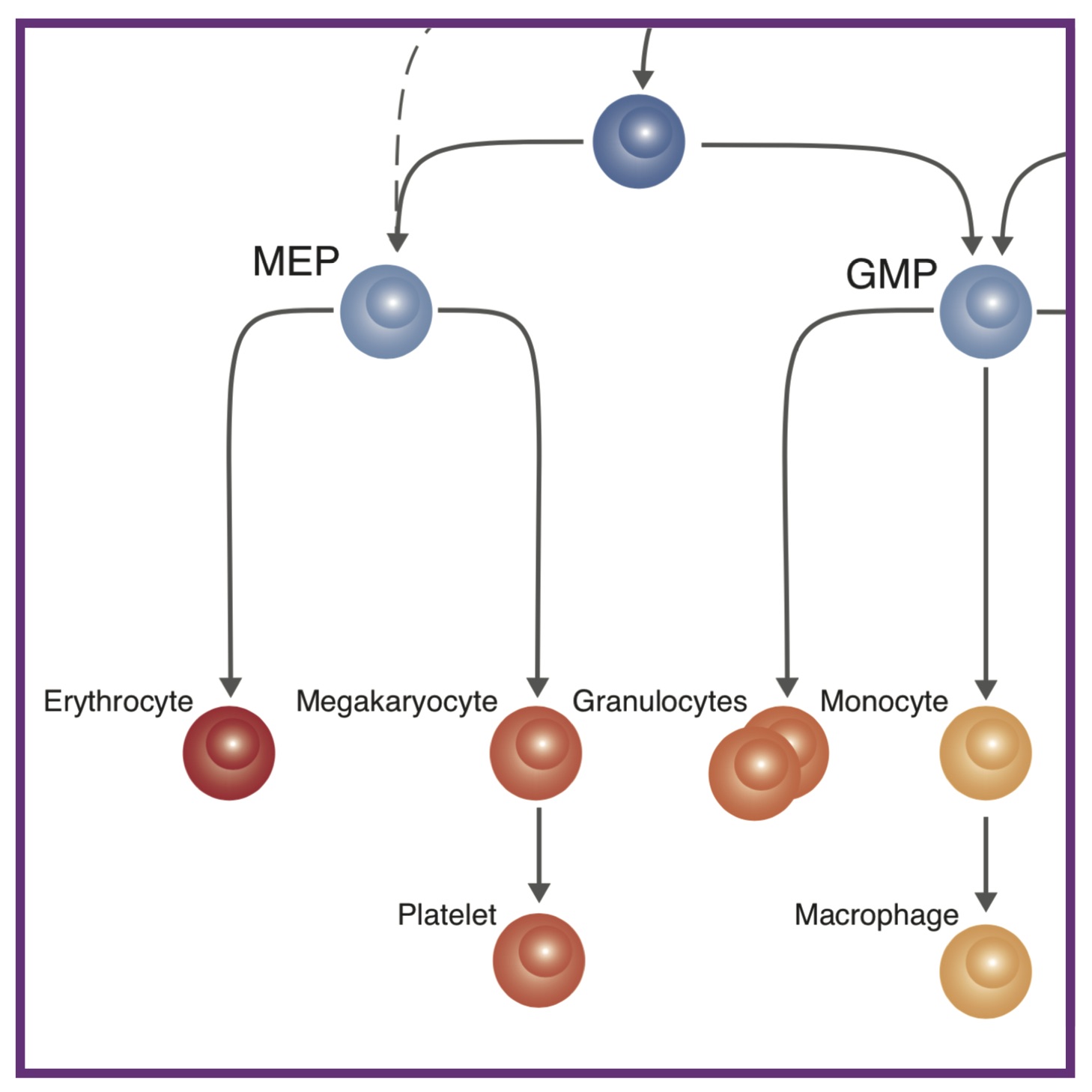
 11. Single Cell Phenotyping Reveals Heterogeneity among Haematopoietic Stem Cells
11. Single Cell Phenotyping Reveals Heterogeneity among Haematopoietic Stem CellsAL MacLean*, MA Smith*, J Liepe, A Sim, R Khorshed, NM Rashidi, N Scherf, A Krinner, I Roeder, C Lo Celso, MPH Stumpf
AL MacLean*, MA Smith*, J Liepe, et al.
Stem Cells, 2017. 35(11):2292-2304 Pubmed Stem Cells
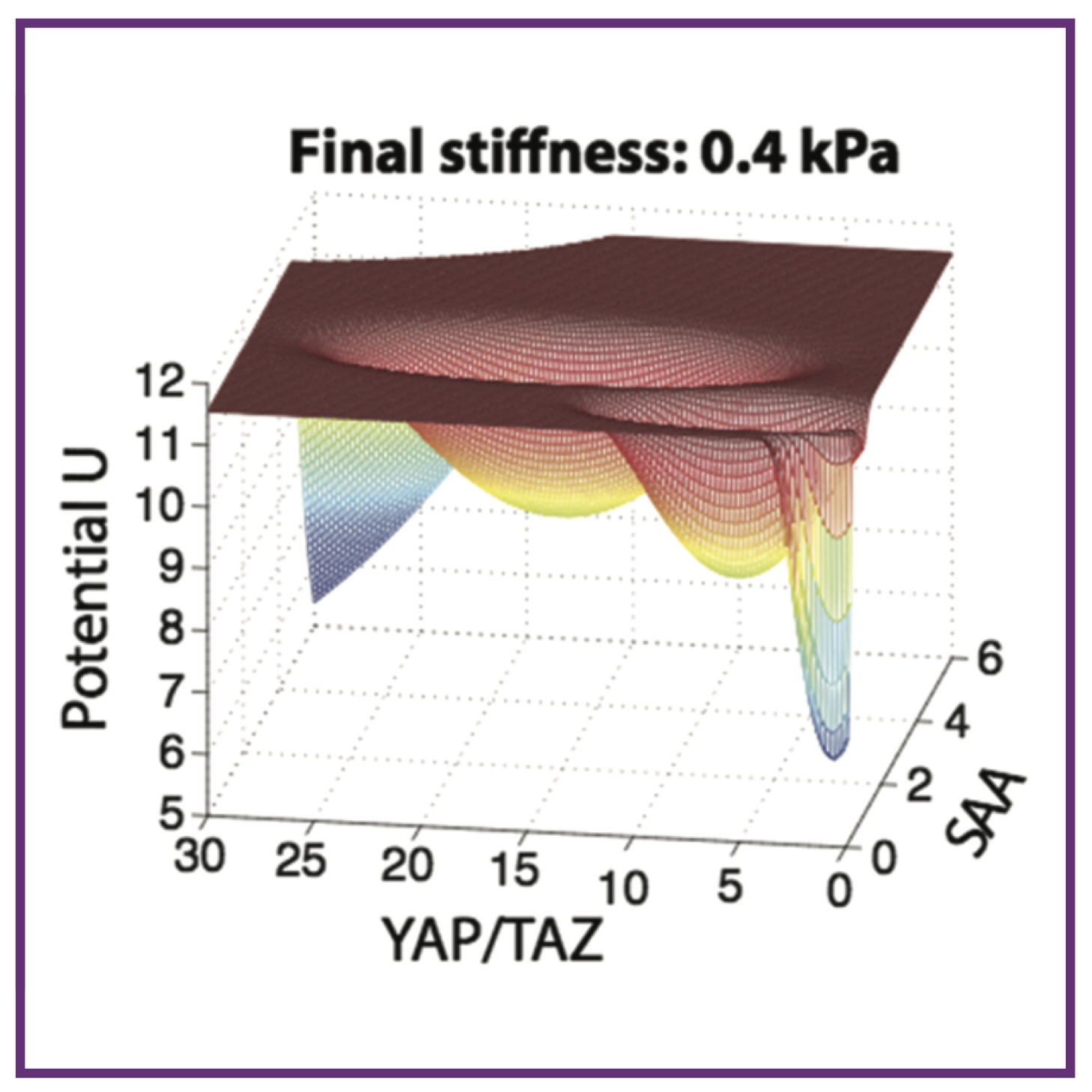 A mathematical model of mechanotransduction reveals how mechanical memory regulates mesenchymal stem cell fate decisions
A mathematical model of mechanotransduction reveals how mechanical memory regulates mesenchymal stem cell fate decisionsT Peng*, L Liu*, AL MacLean, CW Wong, W Zhao, Q Nie
T Peng*, L Liu*, AL MacLean, et al.
BMC Sys Bio, 2017. 11(1):55 Pubmed BMC Syst Biol
AL MacLean, C Lo Celso, MPH Stumpf
Stem Cells, 2017. 35(1):80-88 Pubmed Stem Cells
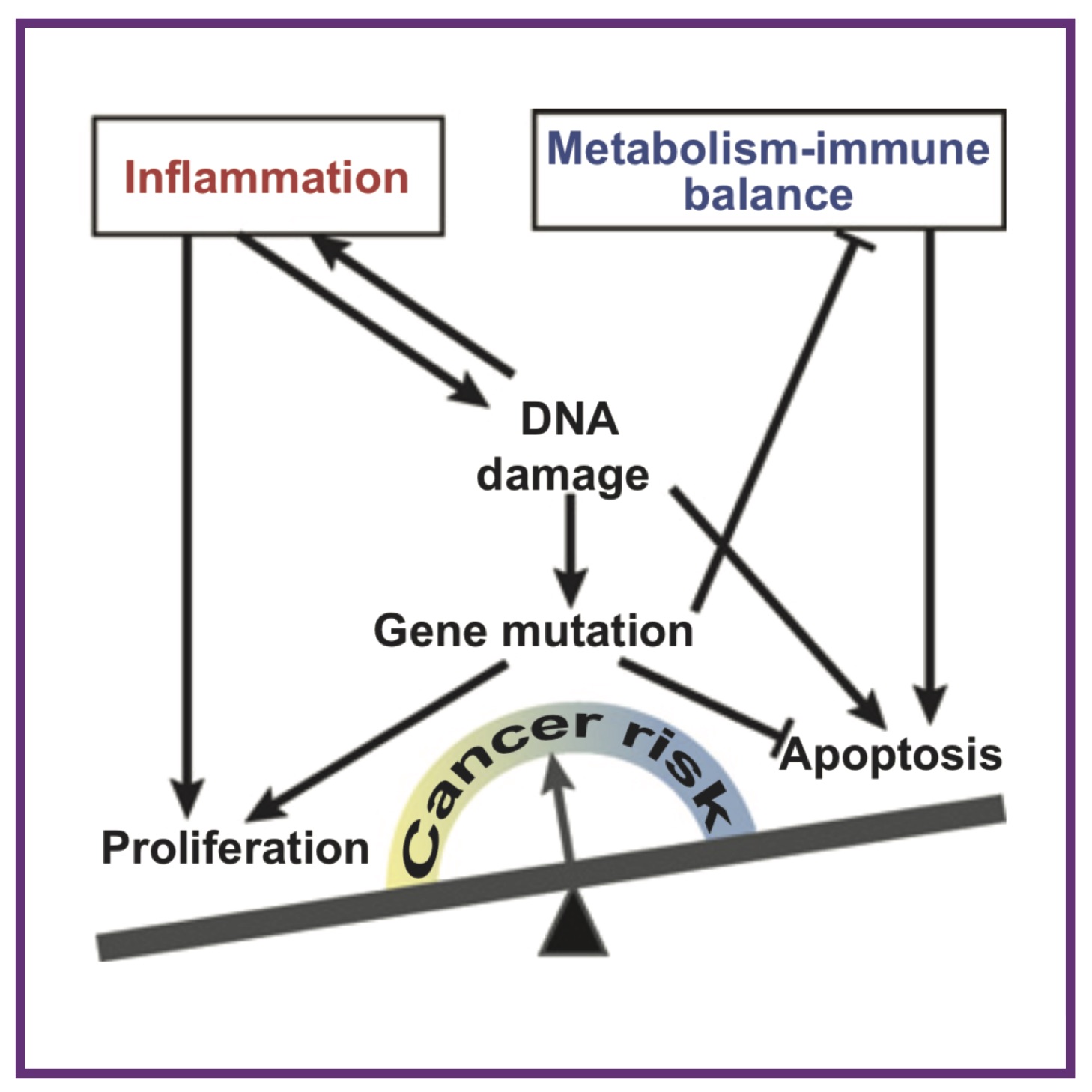
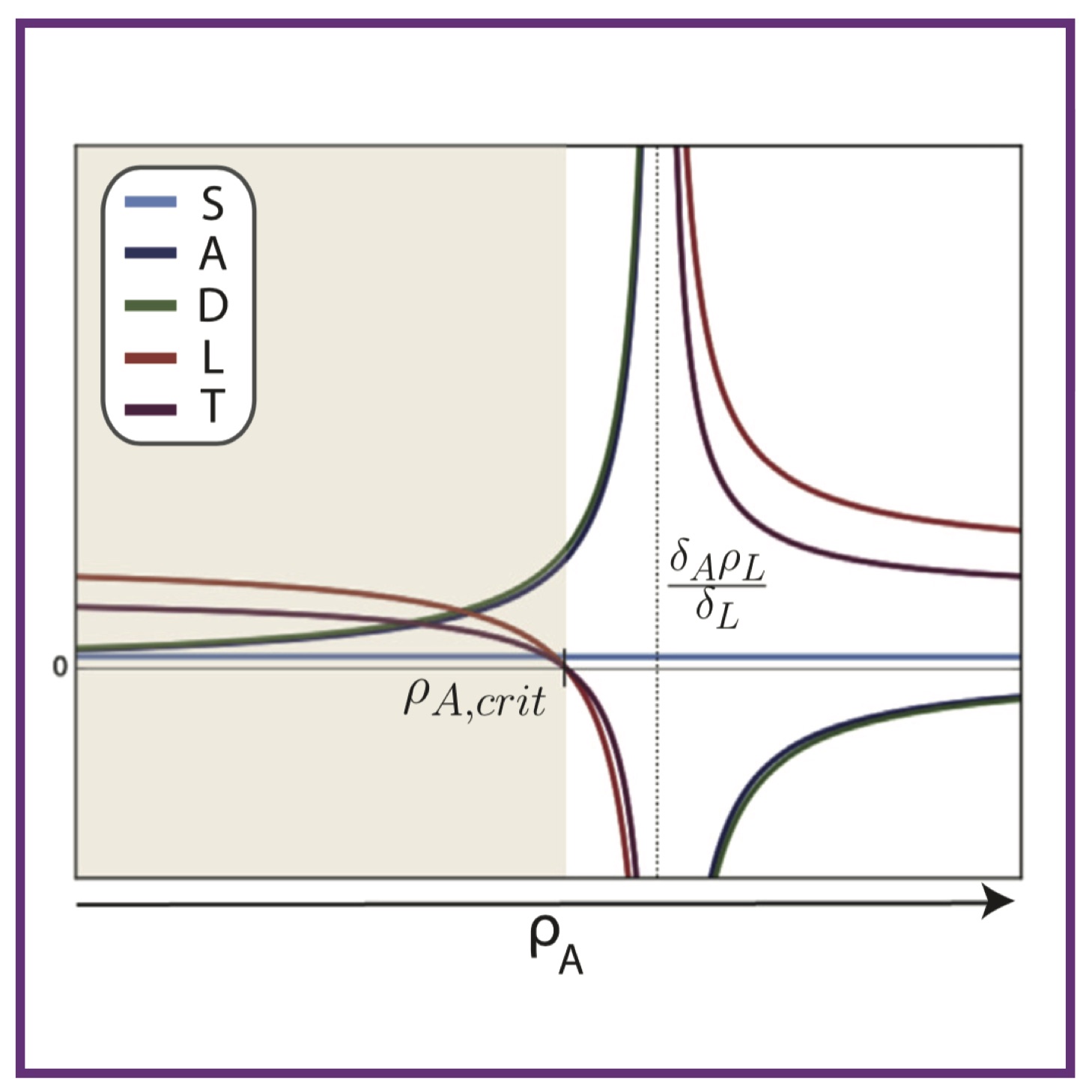 Feedback mechanisms control coexistence in a stem cell model of acute myeloid leukaemia
Feedback mechanisms control coexistence in a stem cell model of acute myeloid leukaemiaHL Crowell*, AL MacLean*, MPH Stumpf
J Theor Biol, 2016. 401:43-53 Pubmed J Theor Biol
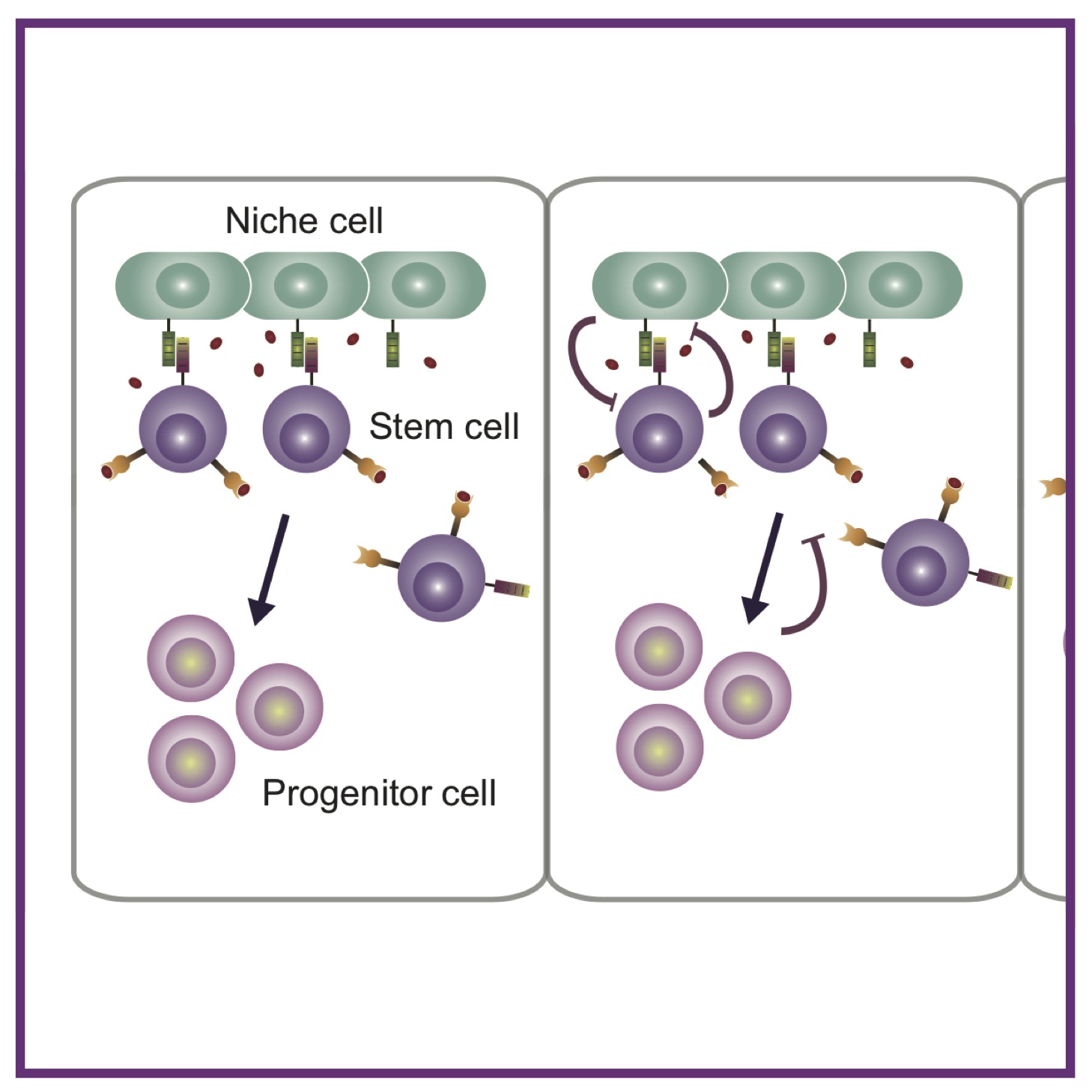
AL MacLean*, P Kirk*, MPH Stumpf
Biol Open, 2015. 4:1420-1426 Pubmed Biol Open
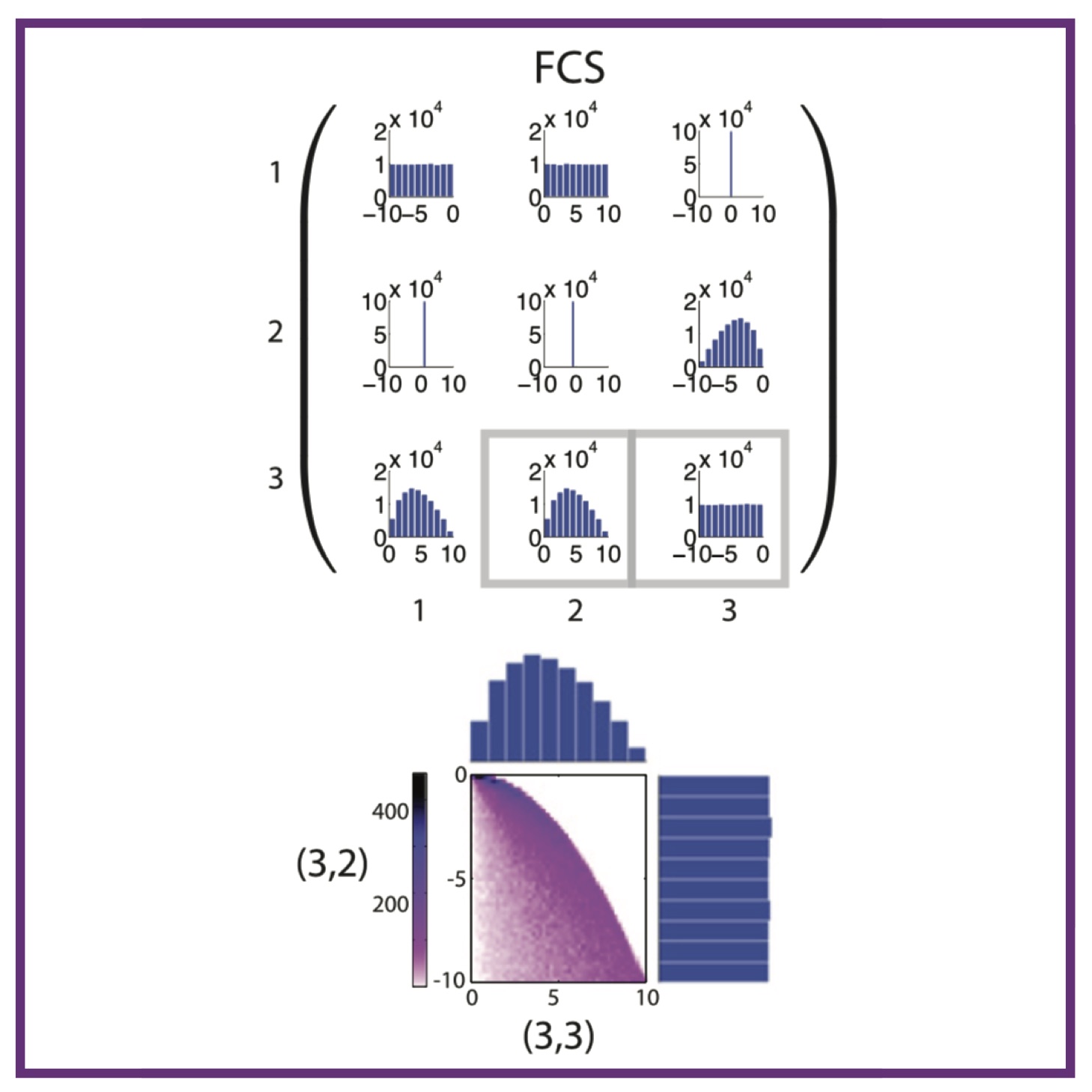
P Kirk*, DMY Rolando*, AL MacLean, MPH Stumpf
New J Phys, 2015, 17(8) New J Phys
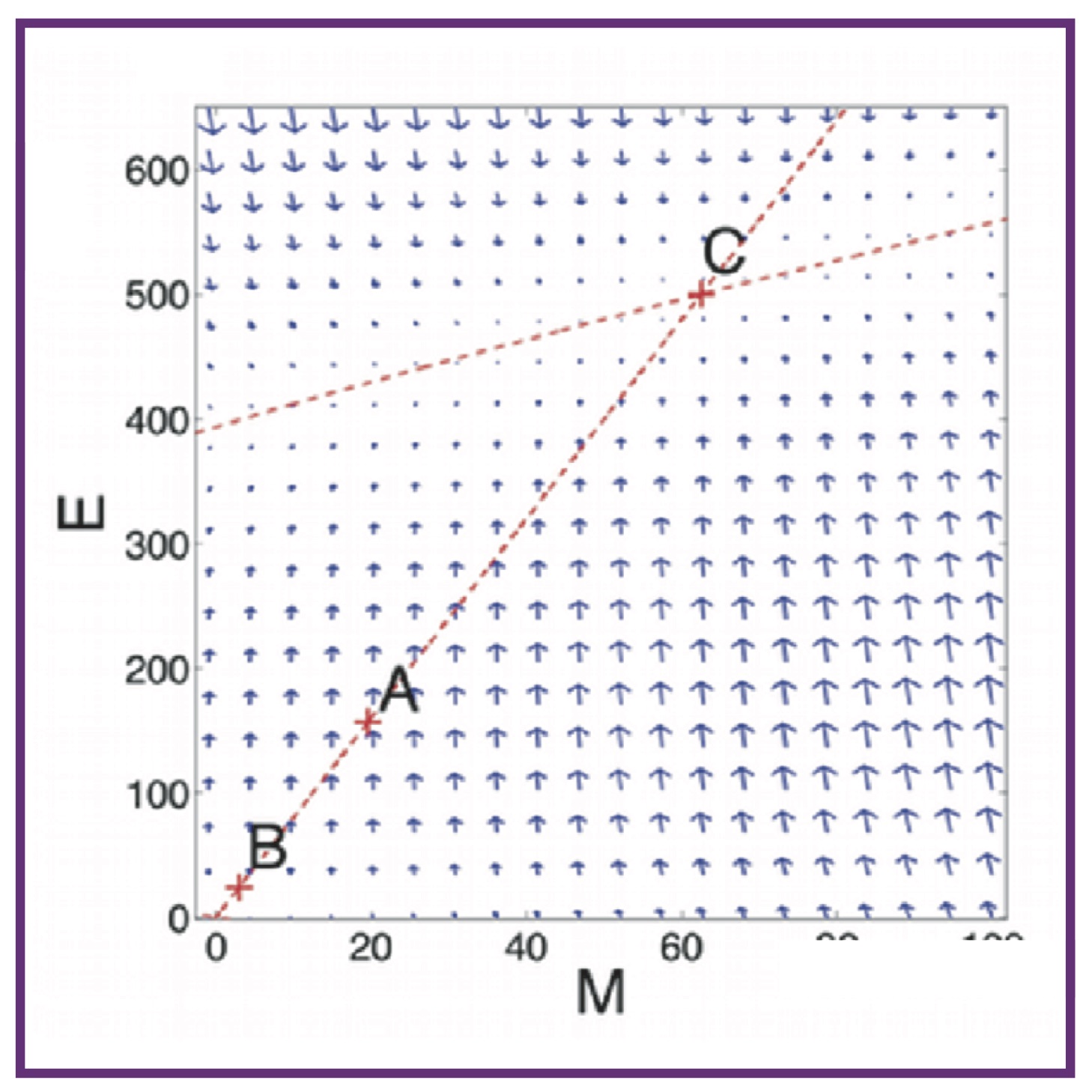 Epithelial-mesenchymal transition in metastases affects tumor dormancy in a simple mathematical model
Epithelial-mesenchymal transition in metastases affects tumor dormancy in a simple mathematical modelAL MacLean, HA Harrington, MPH Stumpf, MDH Hansen
Biomedicines, 2014. 2(4):384-402 Pubmed Biomedicines
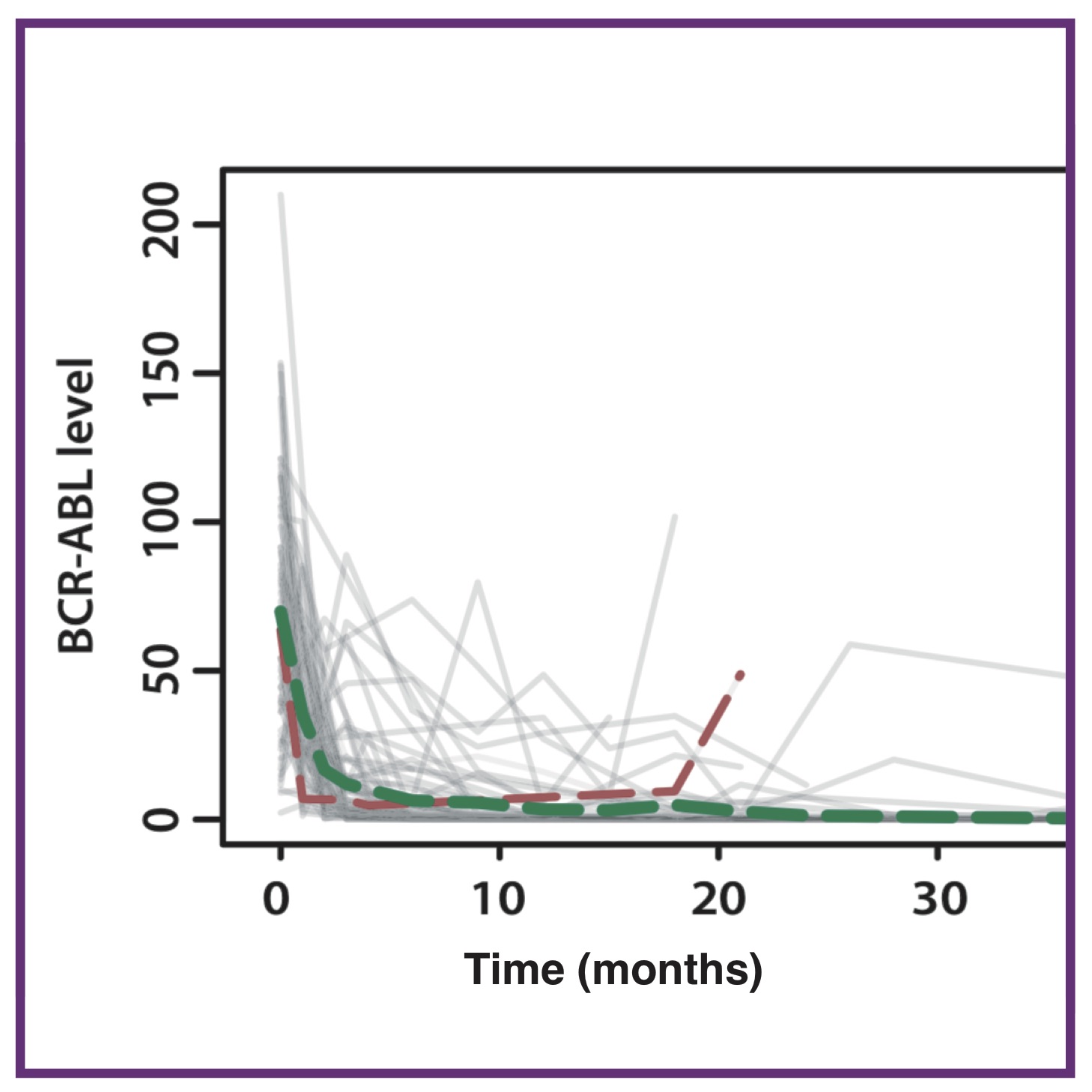 Ecology in the hematopoietic stem cell niche determines the clinical outcome in chronic myeloid leukemia
Ecology in the hematopoietic stem cell niche determines the clinical outcome in chronic myeloid leukemiaAL MacLean*, S Filippi*, MPH Stumpf
Proc Natl Acad Sci USA, 2014. 111(10):3882-88 Pubmed PNAS
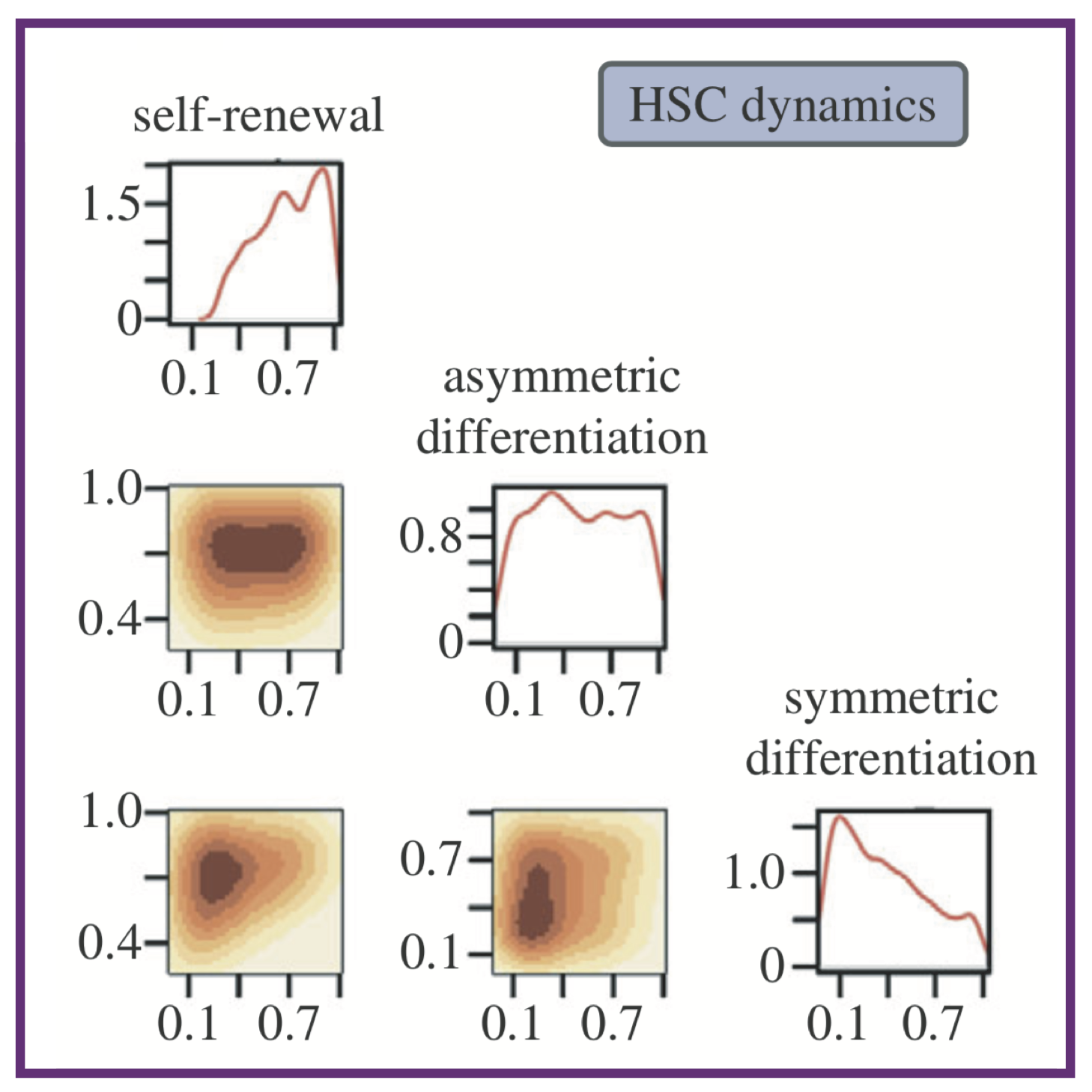 Population dynamics of normal and leukaemia stem cells in the haematopoietic stem cell niche show distinct regimes where leukaemia will be controlled
Population dynamics of normal and leukaemia stem cells in the haematopoietic stem cell niche show distinct regimes where leukaemia will be controlledAL MacLean, C Lo Celso, MPH Stumpf
J R Soc Interface, 2013. 10(81):20120968 Pubmed J R Soc Interface
Peer-reviewed Book Chapters
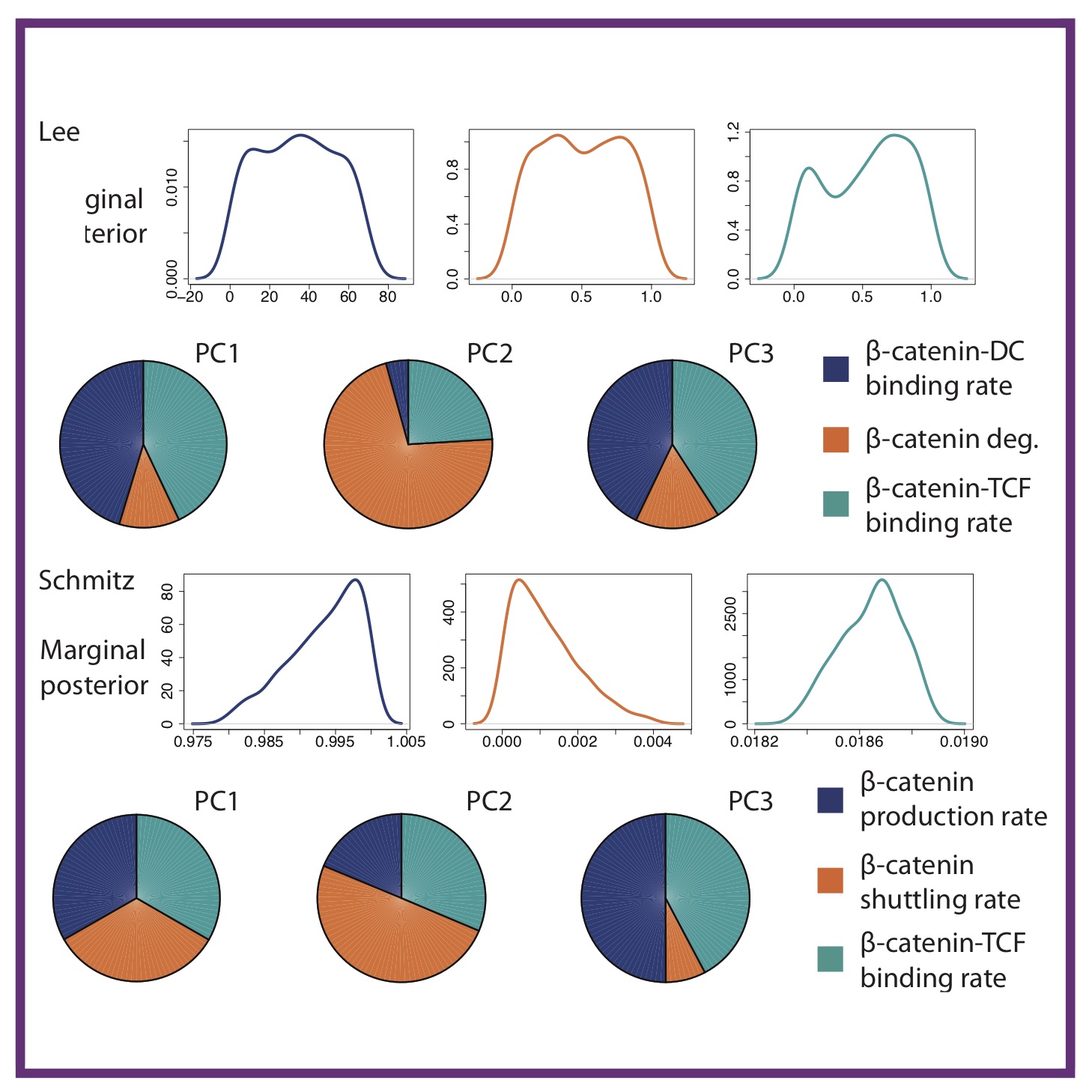 Mathematical and Statistical Techniques for Systems Medicine: The Wnt Signaling Pathway as a Case Study
Mathematical and Statistical Techniques for Systems Medicine: The Wnt Signaling Pathway as a Case StudyAL MacLean, HA Harrington, MPH Stumpf, HM Byrne
AL MacLean, HA Harrington, MPH Stumpf, et al.
Systems Medicine: Meth Molecular Biology: Springer, 2016 Springer Chapter


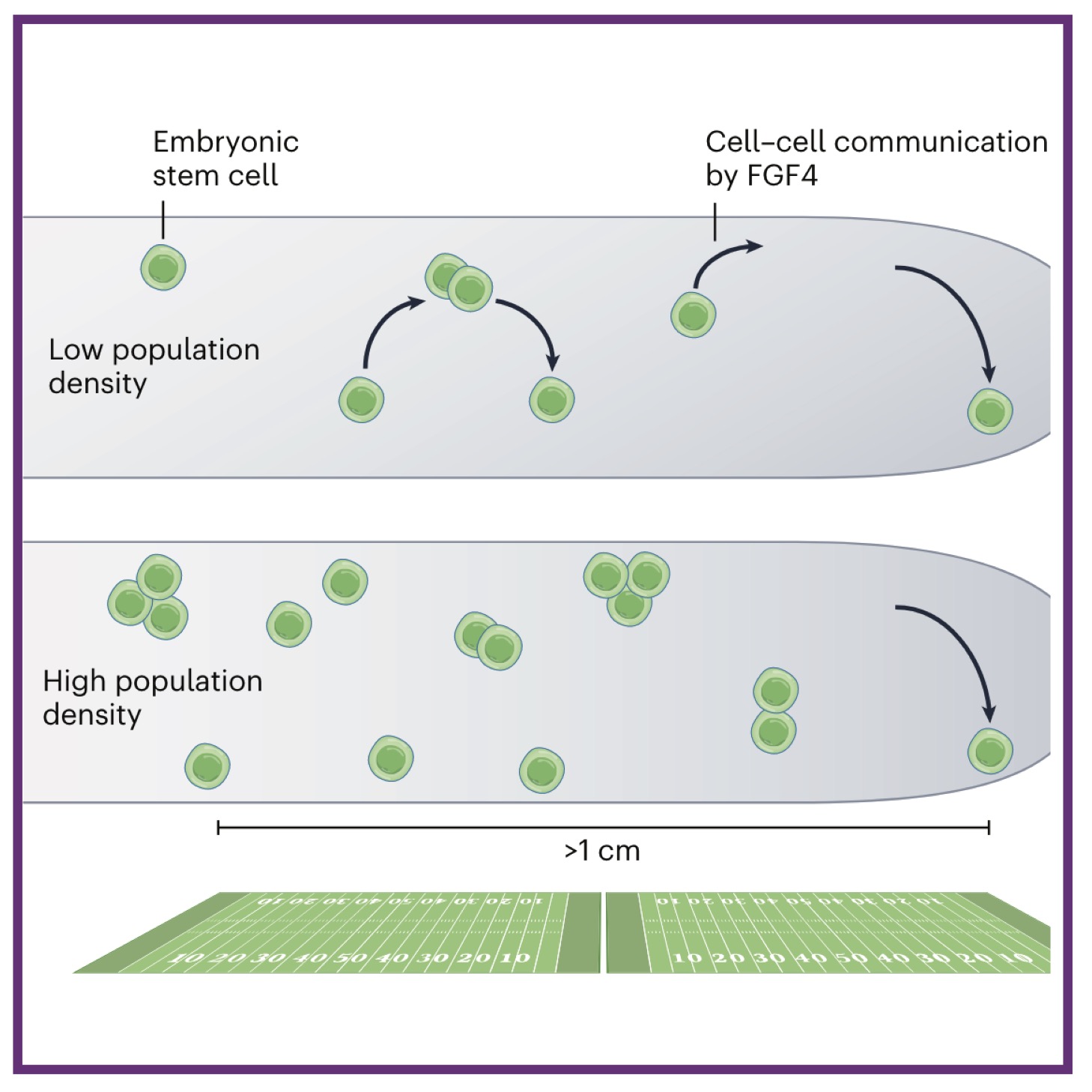
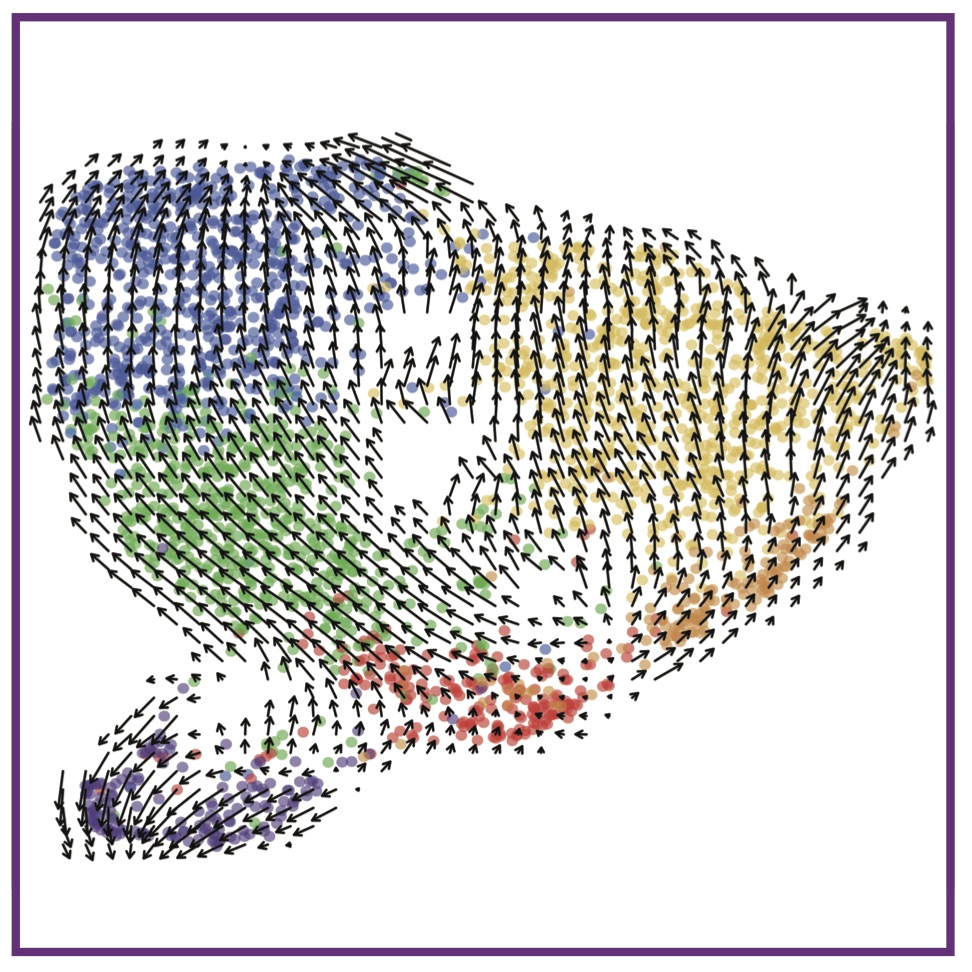 Defining epidermal basal cell states during skin homeostasis and wound healing using single-cell transcriptomics
Defining epidermal basal cell states during skin homeostasis and wound healing using single-cell transcriptomics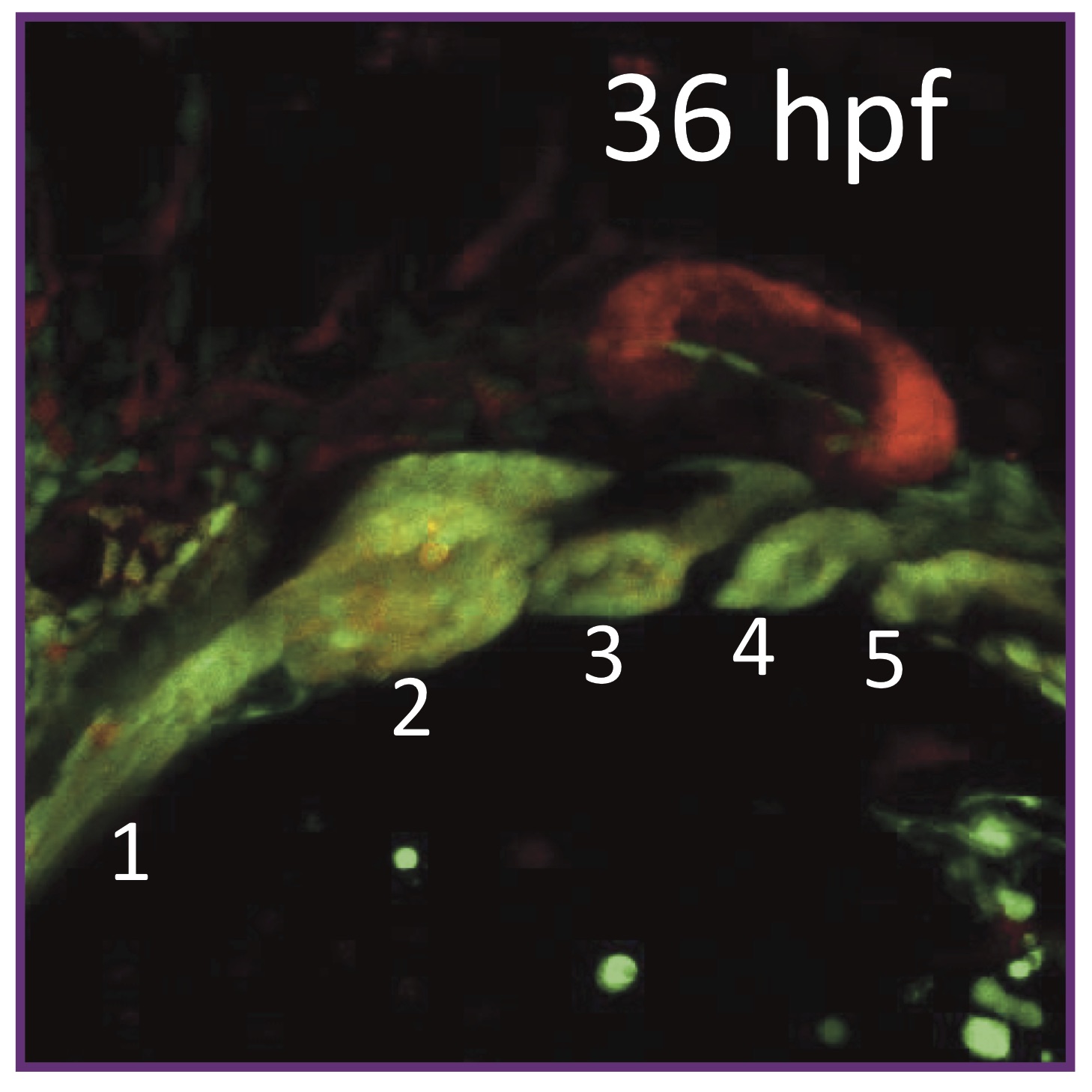 Transcriptomics reveals complex kinetics of dorsal‐ventral patterning gene expression in the mandibular arch
Transcriptomics reveals complex kinetics of dorsal‐ventral patterning gene expression in the mandibular arch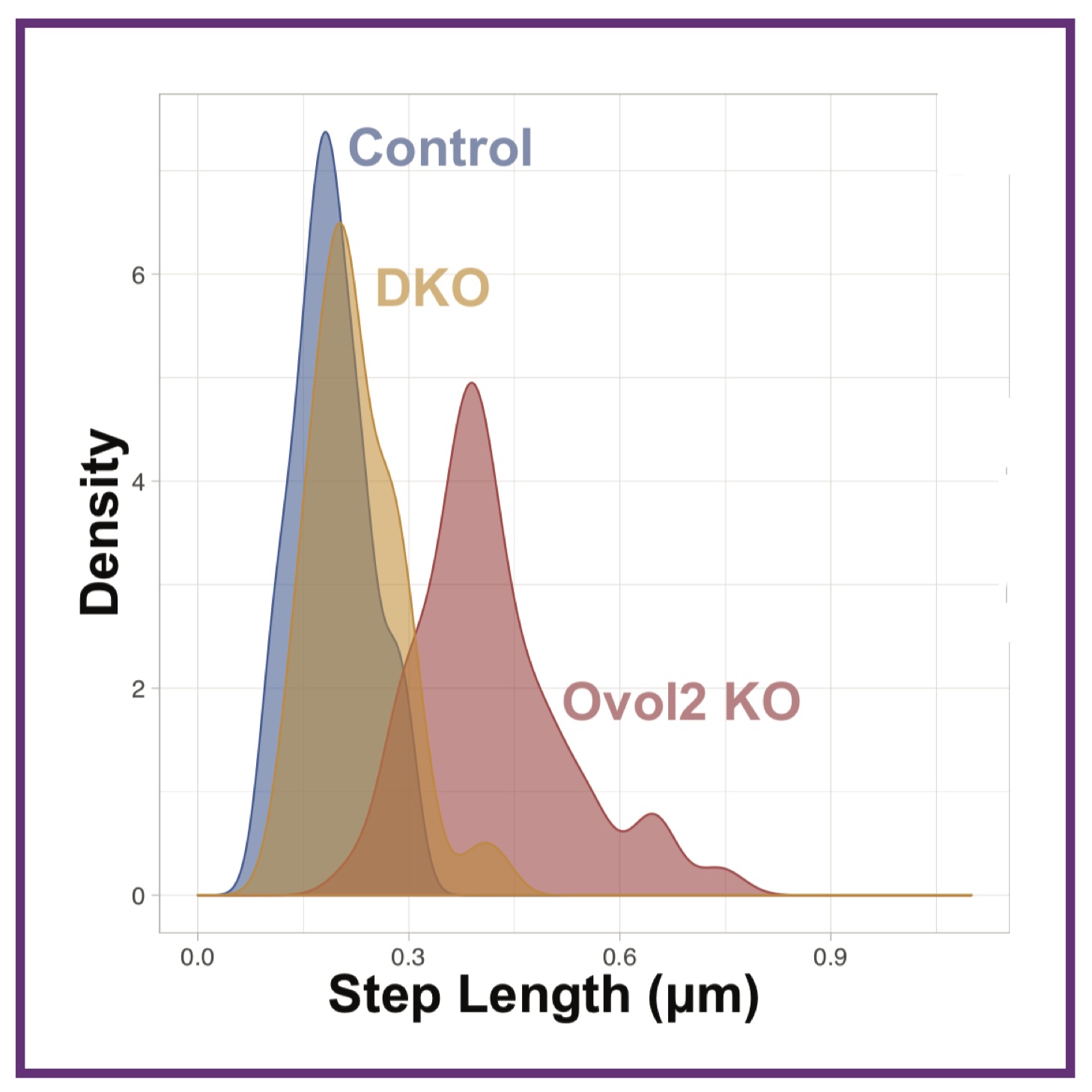 An Ovol2‐Zeb1 transcriptional circuit regulates epithelial directional migration and proliferation
An Ovol2‐Zeb1 transcriptional circuit regulates epithelial directional migration and proliferation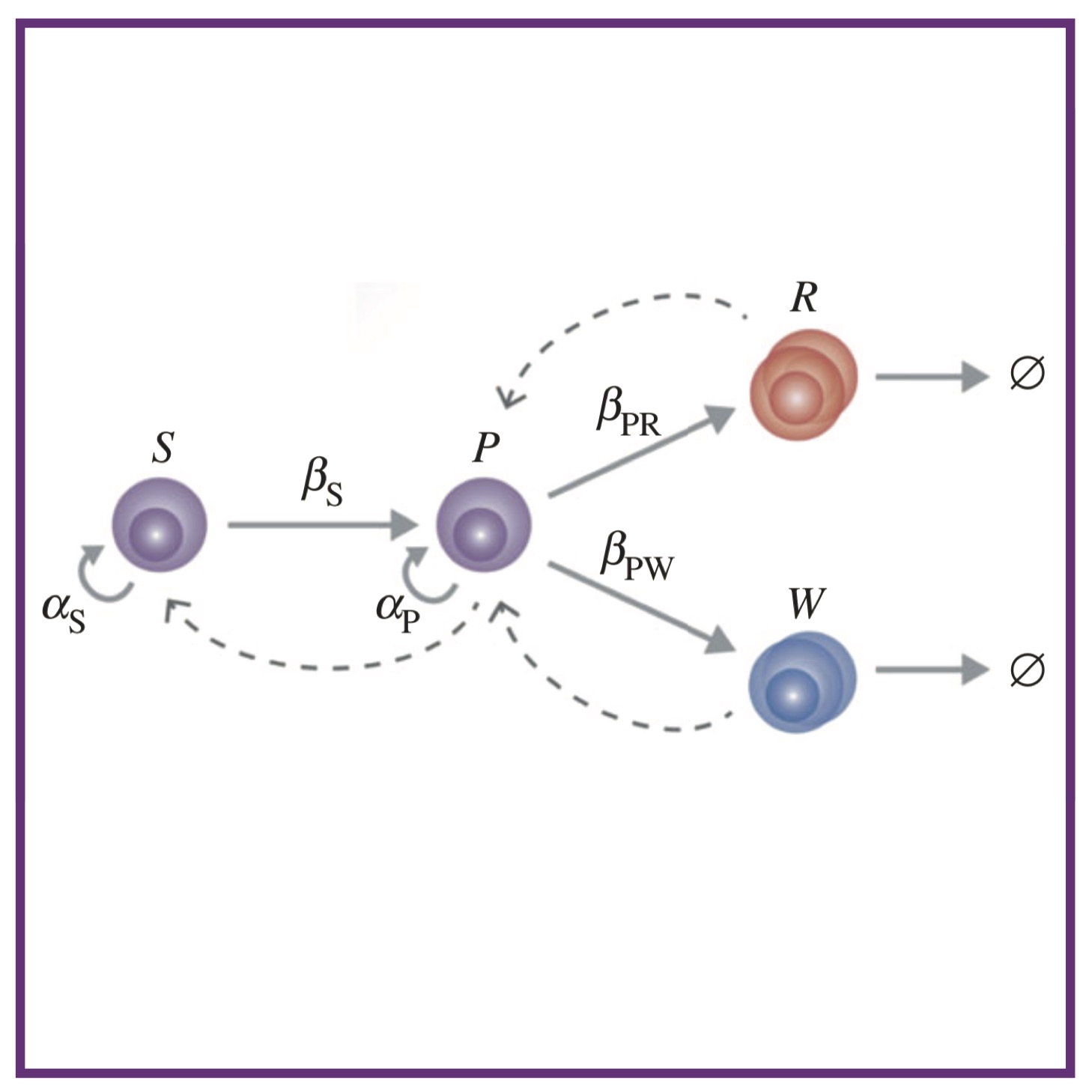 Systematic tracking of altered haematopoiesis during sporozoite-mediated malaria development reveals multiple response points
Systematic tracking of altered haematopoiesis during sporozoite-mediated malaria development reveals multiple response points Parameter-free methods distinguish Wnt pathway models and guide design of experiments
Parameter-free methods distinguish Wnt pathway models and guide design of experiments